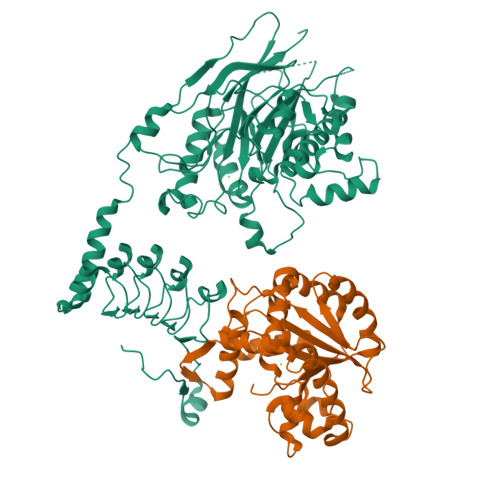
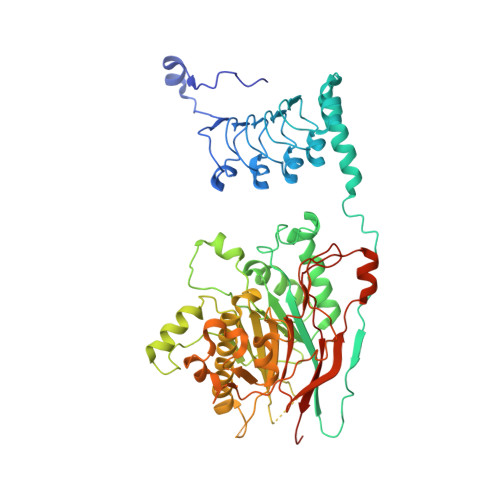
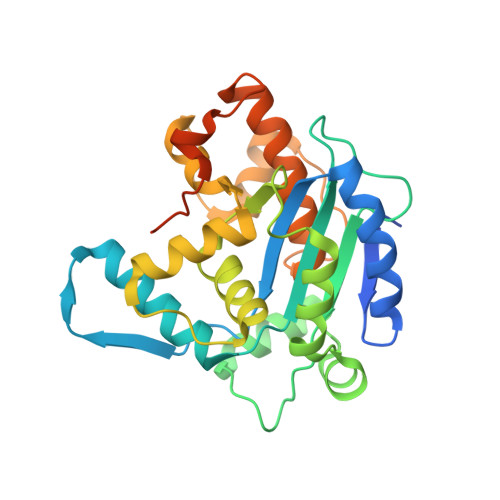
Crystal structure and functional properties of the human CCR4-CAF1 deadenylase complex.
Chen, Y., Khazina, E., Izaurralde, E., Weichenrieder, O.(2021) Nucleic Acids Res 49: 6489-6510
- PubMed: 34038562
- DOI: https://doi.org/10.1093/nar/gkab414
- Primary Citation of Related Structures:
7AX1 - PubMed Abstract:
The CCR4 and CAF1 deadenylases physically interact to form the CCR4-CAF1 complex and function as the catalytic core of the larger CCR4-NOT complex. Together, they are responsible for the eventual removal of the 3'-poly(A) tail from essentially all cellular mRNAs and consequently play a central role in the posttranscriptional regulation of gene expression. The individual properties of CCR4 and CAF1, however, and their respective contributions in different organisms and cellular environments are incompletely understood. Here, we determined the crystal structure of a human CCR4-CAF1 complex and characterized its enzymatic and substrate recognition properties. The structure reveals specific molecular details affecting RNA binding and hydrolysis, and confirms the CCR4 nuclease domain to be tethered flexibly with a considerable distance between both enzyme active sites. CCR4 and CAF1 sense nucleotide identity on both sides of the 3'-terminal phosphate, efficiently differentiating between single and consecutive non-A residues. In comparison to CCR4, CAF1 emerges as a surprisingly tunable enzyme, highly sensitive to pH, magnesium and zinc ions, and possibly allowing distinct reaction geometries. Our results support a picture of CAF1 as a primordial deadenylase, which gets assisted by CCR4 for better efficiency and by the assembled NOT proteins for selective mRNA targeting and regulation.
Organizational Affiliation:
Department of Biochemistry, Max Planck Institute for Developmental Biology, Max-Planck-Ring 5, D-72076 Tübingen, Germany.