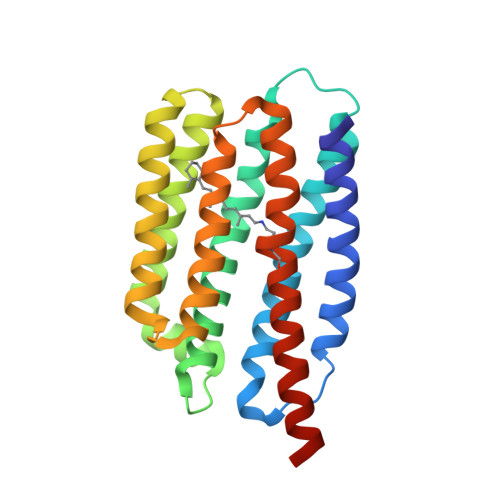
Viral rhodopsins 1 are an unique family of light-gated cation channels.
Zabelskii, D., Alekseev, A., Kovalev, K., Rankovic, V., Balandin, T., Soloviov, D., Bratanov, D., Savelyeva, E., Podolyak, E., Volkov, D., Vaganova, S., Astashkin, R., Chizhov, I., Yutin, N., Rulev, M., Popov, A., Eria-Oliveira, A.S., Rokitskaya, T., Mager, T., Antonenko, Y., Rosselli, R., Armeev, G., Shaitan, K., Vivaudou, M., Buldt, G., Rogachev, A., Rodriguez-Valera, F., Kirpichnikov, M., Moser, T., Offenhausser, A., Willbold, D., Koonin, E., Bamberg, E., Gordeliy, V.(2020) Nat Commun 11: 5707-5707
- PubMed: 33177509
- DOI: https://doi.org/10.1038/s41467-020-19457-7
- Primary Citation of Related Structures:
7AKW, 7AKX, 7AKY - PubMed Abstract:
Phytoplankton is the base of the marine food chain as well as oxygen and carbon cycles and thus plays a global role in climate and ecology. Nucleocytoplasmic Large DNA Viruses that infect phytoplankton organisms and regulate the phytoplankton dynamics encompass genes of rhodopsins of two distinct families. Here, we present a functional and structural characterization of two proteins of viral rhodopsin group 1, OLPVR1 and VirChR1. Functional analysis of VirChR1 shows that it is a highly selective, Na + /K + -conducting channel and, in contrast to known cation channelrhodopsins, it is impermeable to Ca 2+ ions. We show that, upon illumination, VirChR1 is able to drive neural firing. The 1.4 Å resolution structure of OLPVR1 reveals remarkable differences from the known channelrhodopsins and a unique ion-conducting pathway. Thus, viral rhodopsins 1 represent a unique, large group of light-gated channels (viral channelrhodopsins, VirChR1s). In nature, VirChR1s likely mediate phototaxis of algae enhancing the host anabolic processes to support virus reproduction, and therefore, might play a major role in global phytoplankton dynamics. Moreover, VirChR1s have unique potential for optogenetics as they lack possibly noxious Ca 2+ permeability.
Organizational Affiliation:
Institute of Biological Information Processing (IBI-7: Structural Biochemistry), Forschungszentrum Jülich GmbH, Jülich, Germany.