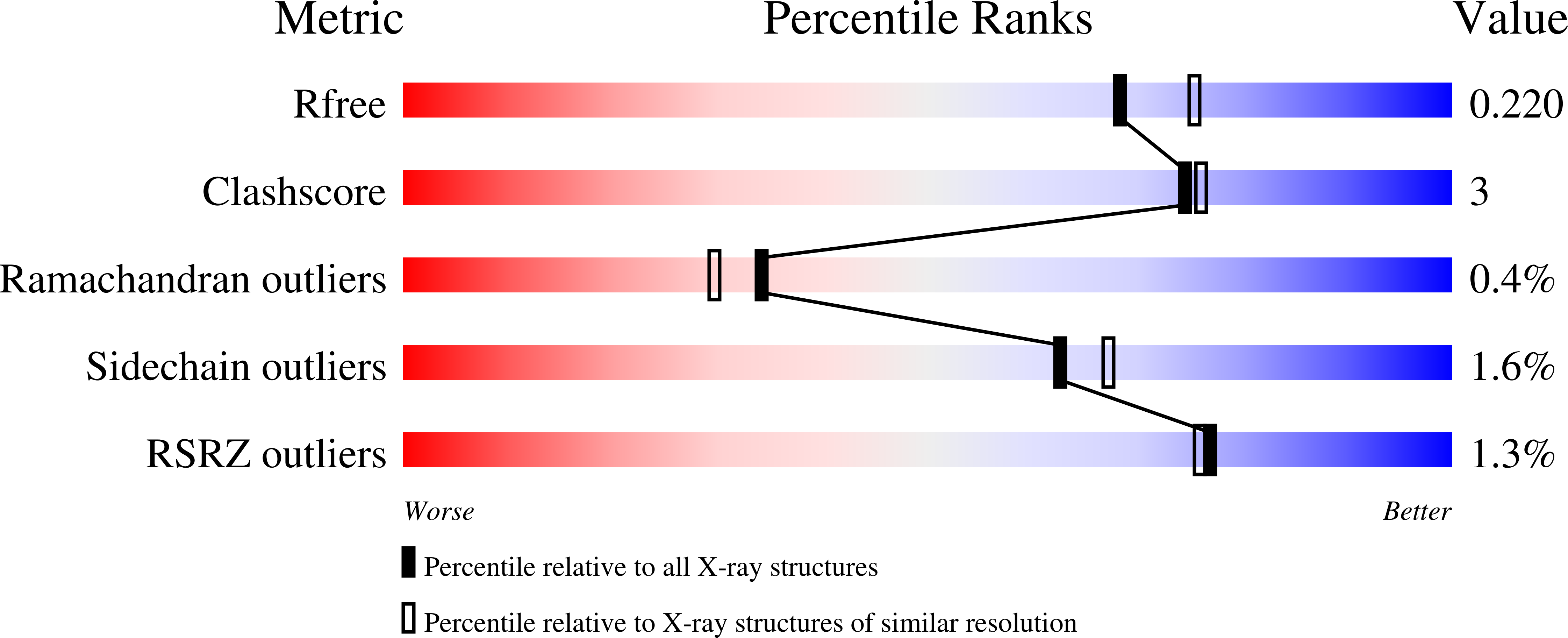
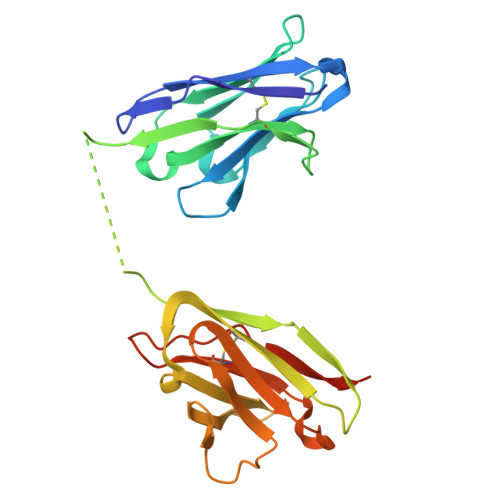
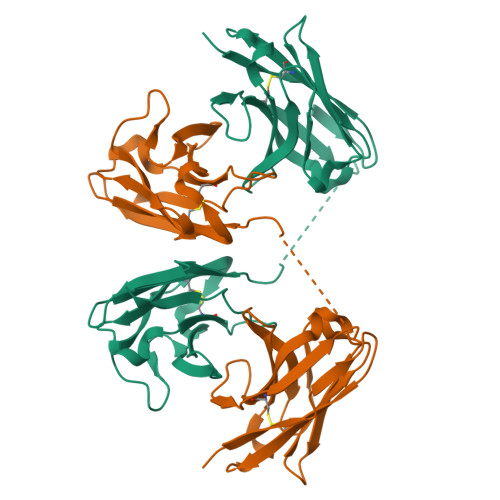
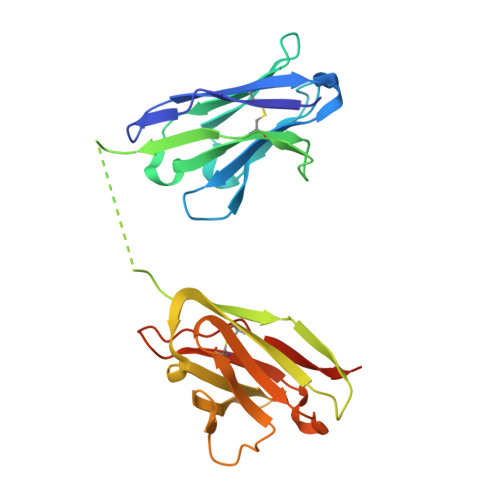
Inference of molecular structure for characterization and improvement of clinical grade immunocytokines.
Ongaro, T., Guarino, S.R., Scietti, L., Palamini, M., Wulhfard, S., Neri, D., Villa, A., Forneris, F.(2021) J Struct Biol 213: 107696-107696
- PubMed: 33493635
- DOI: https://doi.org/10.1016/j.jsb.2021.107696
- Primary Citation of Related Structures:
7AH1 - PubMed Abstract:
The use of immunomodulatory agents for the treatment of cancer is gaining a growing biopharmaceutical interest. Antibody-cytokine fusion proteins, namely immunocytokines, represent a promising solution for the regulation of the immune system at the site of disease. The three-dimensional arrangement of these molecules can profoundly influence their biological activity and pharmacokinetic properties. Structural techniques might provide important insight in the 3D arrangement of immunocytokines. Here, we performed structure investigations on clinical grade fusion proteins L19-IL2, IL12-L19L19 and L19L19-IL2 to elucidate their quaternary organization. Crystallographic characterization of the common L19 antibody fragment at a resolution of 2.0-Å was combined with low-resolution studies of the full-length chimeric molecules using small-angle synchrotron X-ray scattering (SAXS) and negative stain electron microscopy. Characterization of the full-length quaternary structures of the immunocytokines in solution by SAXS consistently supported the diabody structure in the L19-IL2 immunocytokine and allowed generation of low-resolution models of the chimeric proteins L19L19-IL2 and IL12-L19L19. Comparison with 3D reconstructions obtained from negative-stain electron microscopy revealed marked flexibility associated to the linker regions connecting the cytokine and the antibody components of the chimeric proteins. Collectively, our results indicate that low-resolution molecular structure characterizations provide useful complementary insights for the quality control of immunocytokines, constituting a powerful tool to guide the design and the subsequent optimization steps towards clinical enhancement of these chimeric protein reagents.
Organizational Affiliation:
The Armenise-Harvard Laboratory of Structural Biology, Dept. Biology and Biotechnology, University of Pavia, Via Ferrata 9/A, 27100 Pavia Italy; Philochem AG, Libernstrasse 3, 8112 Otelfingen, Switzerland.