Effect of X-ray free-electron laser-induced shockwaves on haemoglobin microcrystals delivered in a liquid jet.
Grunbein, M.L., Gorel, A., Foucar, L., Carbajo, S., Colocho, W., Gilevich, S., Hartmann, E., Hilpert, M., Hunter, M., Kloos, M., Koglin, J.E., Lane, T.J., Lewandowski, J., Lutman, A., Nass, K., Nass Kovacs, G., Roome, C.M., Sheppard, J., Shoeman, R.L., Stricker, M., van Driel, T., Vetter, S., Doak, R.B., Boutet, S., Aquila, A., Decker, F.J., Barends, T.R.M., Stan, C.A., Schlichting, I.(2021) Nat Commun 12: 1672-1672
- PubMed: 33723266
- DOI: https://doi.org/10.1038/s41467-021-21819-8
- Primary Citation of Related Structures:
7AET, 7AEU, 7AEV - PubMed Abstract:
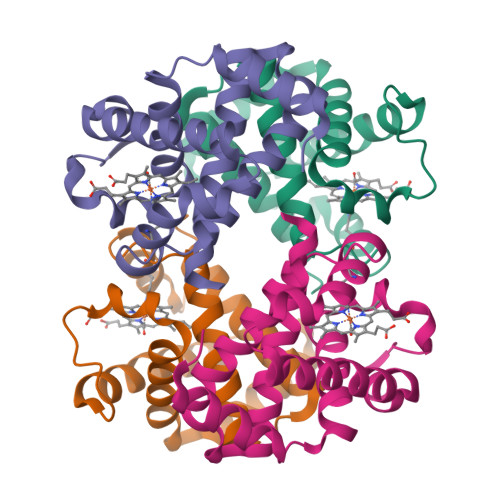
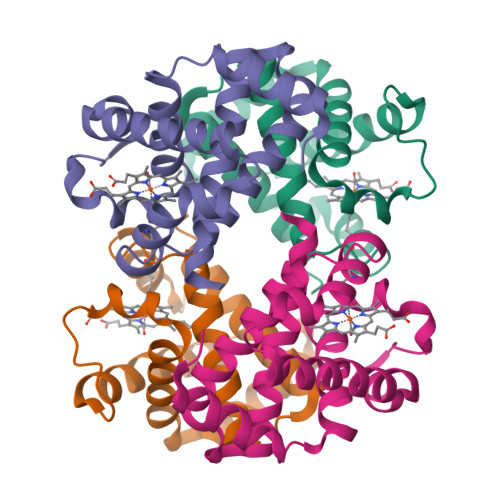
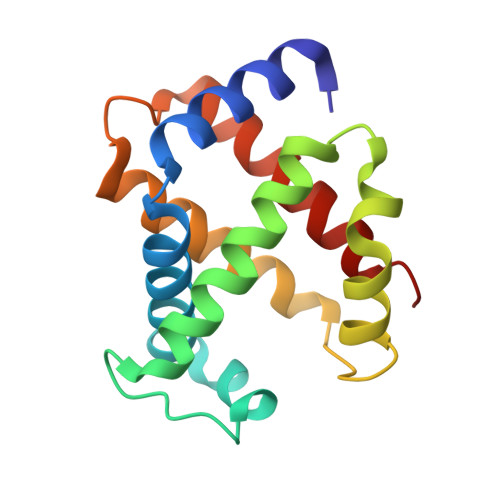
X-ray free-electron lasers (XFELs) enable obtaining novel insights in structural biology. The recently available MHz repetition rate XFELs allow full data sets to be collected in shorter time and can also decrease sample consumption. However, the microsecond spacing of MHz XFEL pulses raises new challenges, including possible sample damage induced by shock waves that are launched by preceding pulses in the sample-carrying jet. We explored this matter with an X-ray-pump/X-ray-probe experiment employing haemoglobin microcrystals transported via a liquid jet into the XFEL beam. Diffraction data were collected using a shock-wave-free single-pulse scheme as well as the dual-pulse pump-probe scheme. The latter, relative to the former, reveals significant degradation of crystal hit rate, diffraction resolution and data quality. Crystal structures extracted from the two data sets also differ. Since our pump-probe attributes were chosen to emulate EuXFEL operation at its 4.5 MHz maximum pulse rate, this prompts concern about such data collection.
Organizational Affiliation:
Max Planck Institute for Medical Research, Jahnstrasse 29, Heidelberg, Germany.