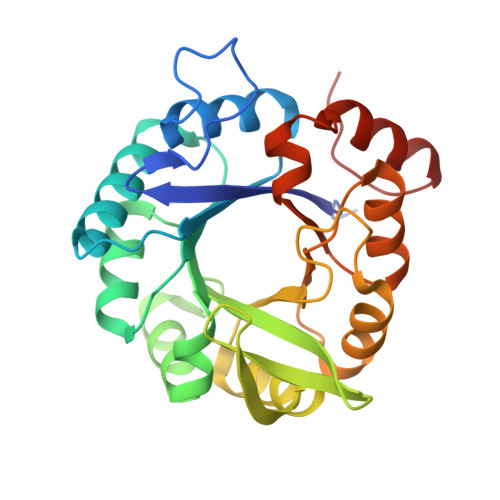
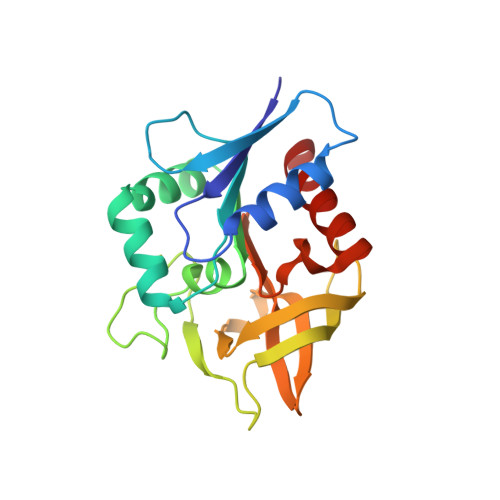
Molecular basis for the allosteric activation mechanism of the heterodimeric imidazole glycerol phosphate synthase complex.
Wurm, J.P., Sung, S., Kneuttinger, A.C., Hupfeld, E., Sterner, R., Wilmanns, M., Sprangers, R.(2021) Nat Commun 12: 2748-2748
- PubMed: 33980881
- DOI: https://doi.org/10.1038/s41467-021-22968-6
- Primary Citation of Related Structures:
7AC8 - PubMed Abstract:
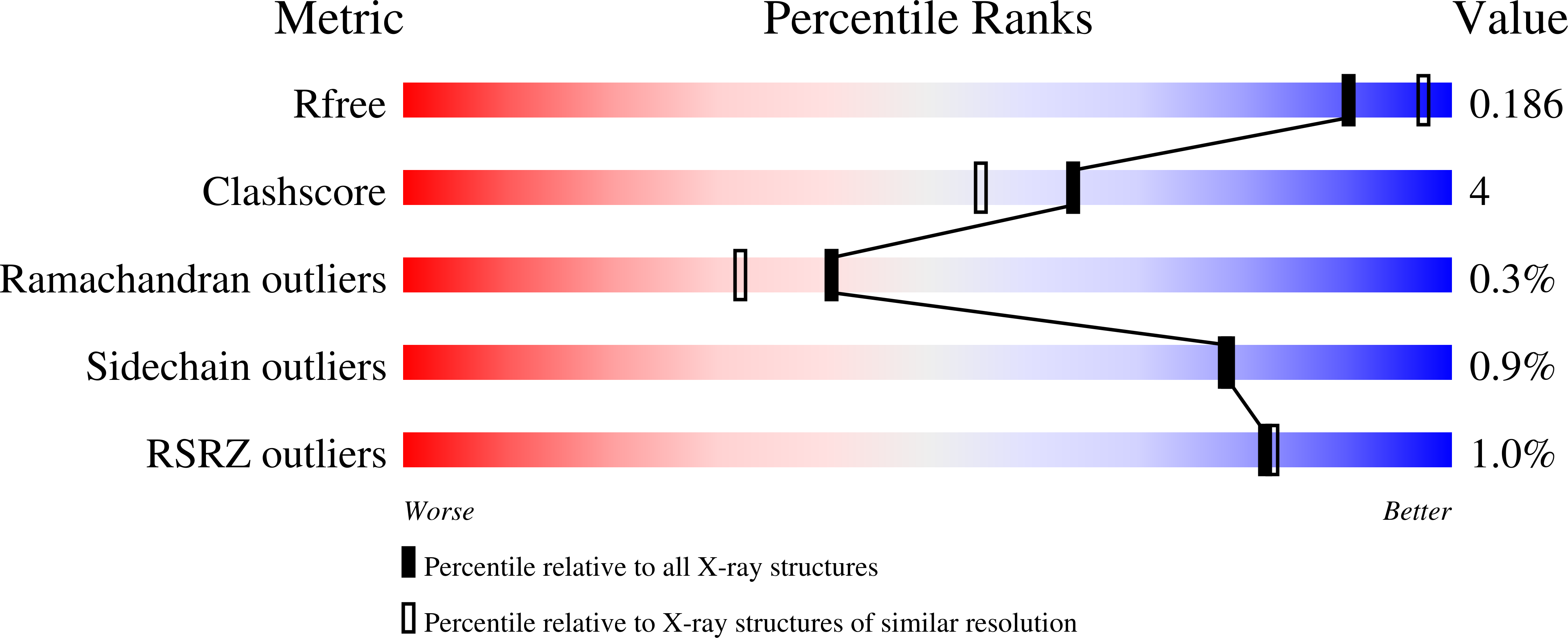
Imidazole glycerol phosphate synthase (HisFH) is a heterodimeric bienzyme complex operating at a central branch point of metabolism. HisFH is responsible for the HisH-catalyzed hydrolysis of glutamine to glutamate and ammonia, which is then used for a cyclase reaction by HisF. The HisFH complex is allosterically regulated but the underlying mechanism is not well understood. Here, we elucidate the molecular basis of the long range, allosteric activation of HisFH. We establish that the catalytically active HisFH conformation is only formed when the substrates of both HisH and HisF are bound. We show that in this conformation an oxyanion hole in the HisH active site is established, which rationalizes the observed 4500-fold allosteric activation compared to the inactive conformation. In solution, the inactive and active conformations are in a dynamic equilibrium and the HisFH turnover rates correlate with the population of the active conformation, which is in accordance with the ensemble model of allostery.
Organizational Affiliation:
Institute of Biophysics and Physical Biochemistry, Regensburg Center for Biochemistry, University of Regensburg, Regensburg, Germany.