PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Manso, J.A., Marcos, T., Ruiz-Martin, V., Casas, J., Alcon, P., Sanchez Crespo, M., Bayon, Y., de Pereda, J.M., Alonso, A.(2022) Cell Mol Life Sci 79: 131-131
- PubMed: 35152348
- DOI: https://doi.org/10.1007/s00018-022-04173-w
- Primary Citation of Related Structures:
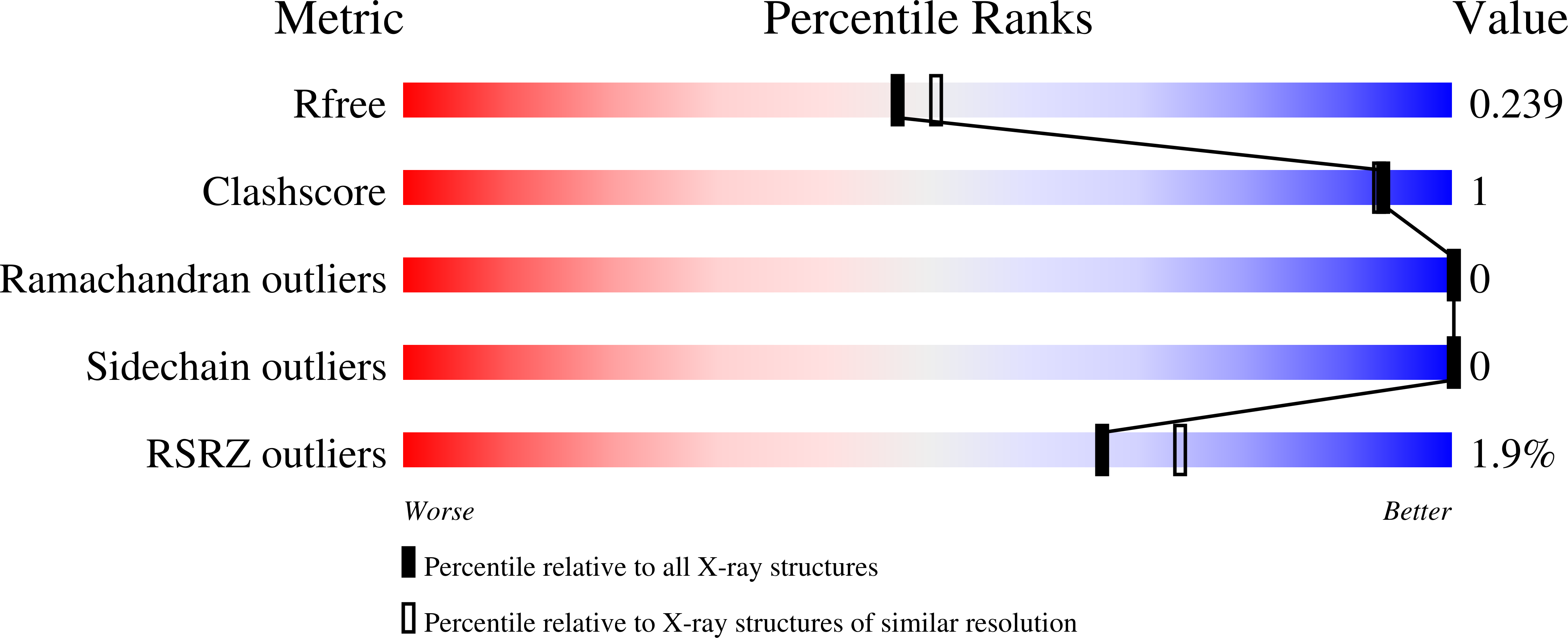
7AAL, 7AAM, 7AAN - PubMed Abstract:
Mutations in the adaptor protein PSTPIP1 cause a spectrum of autoinflammatory diseases, including PAPA and PAMI; however, the mechanism underlying these diseases remains unknown. Most of these mutations lie in PSTPIP1 F-BAR domain, which binds to LYP, a protein tyrosine phosphatase associated with arthritis and lupus. To shed light on the mechanism by which these mutations generate autoinflammatory disorders, we solved the structure of the F-BAR domain of PSTPIP1 alone and bound to the C-terminal homology segment of LYP, revealing a novel mechanism of recognition of Pro-rich motifs by proteins in which a single LYP molecule binds to the PSTPIP1 F-BAR dimer. The residues R228, D246, E250, and E257 of PSTPIP1 that are mutated in immunological diseases directly interact with LYP. These findings link the disruption of the PSTPIP1/LYP interaction to these diseases, and support a critical role for LYP phosphatase in their pathogenesis.
Organizational Affiliation:
Instituto de Biología Molecular y Celular del Cáncer (IBMCC), CSIC-Universidad de Salamanca, Campus Unamuno, 37007, Salamanca, Spain.