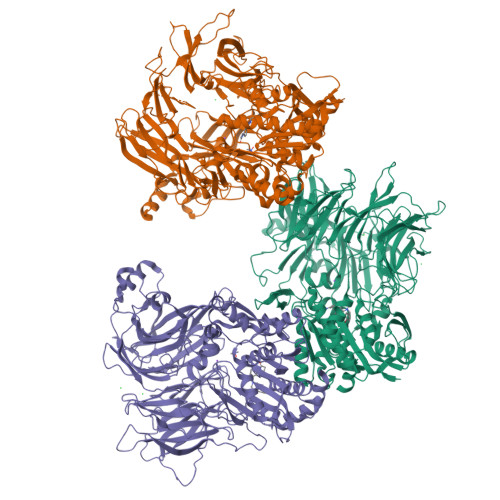
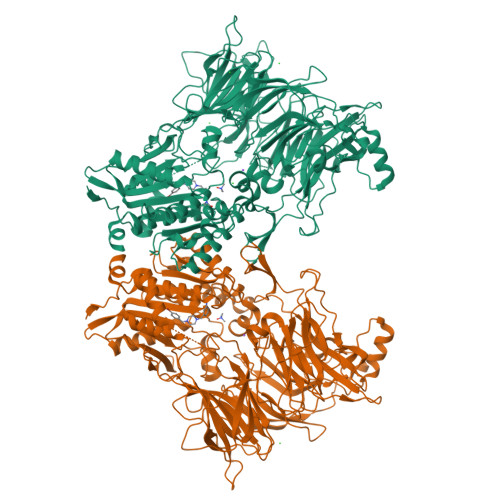
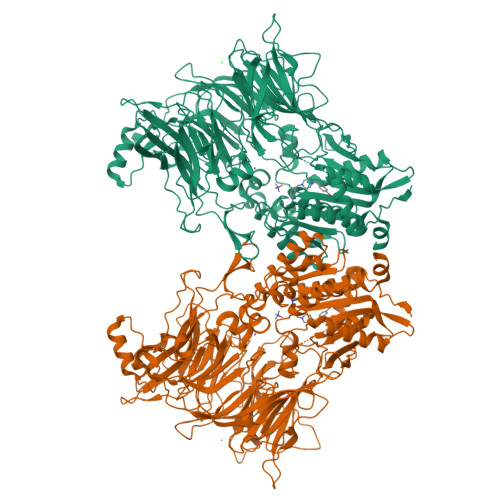
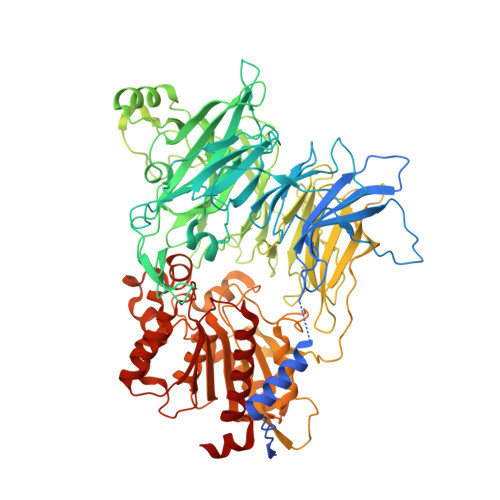
Discovery and Development of 4-Oxo-beta-Lactams as Novel Inhibitors of Dipeptidyl Peptidases 8 and 9
Fehr, L., Carvalho, L.A.R., Ross, B.H., Lum, K., Vieira, A.C., Kiefersauer, R., Geiss-Friedlander, R., Kaiser, M., Rodrigues, T., Lucas, S.D., Cravatt, B.F., Huber, R., Moreira, R.To be published.