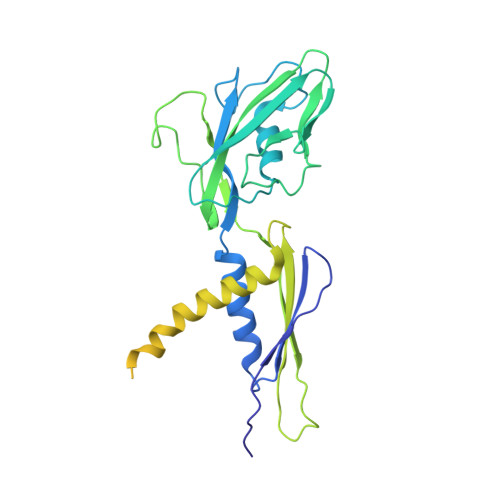
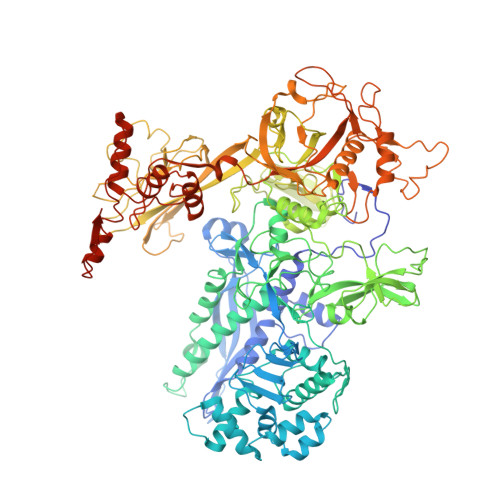
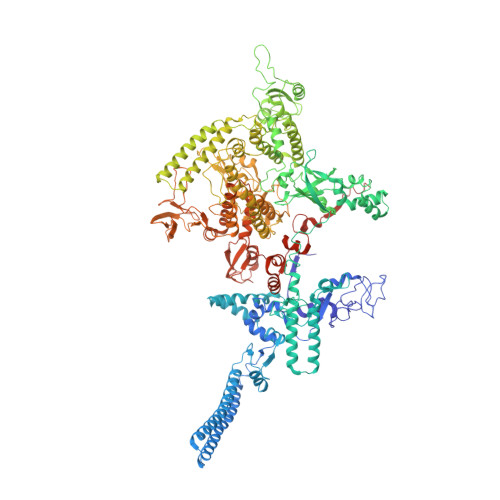
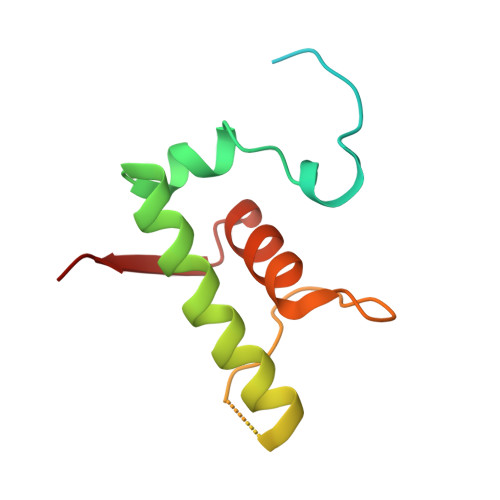
Mycobacterial HelD is a nucleic acids-clearing factor for RNA polymerase.
Kouba, T., Koval', T., Sudzinova, P., Pospisil, J., Brezovska, B., Hnilicova, J., Sanderova, H., Janouskova, M., Sikova, M., Halada, P., Sykora, M., Barvik, I., Novacek, J., Trundova, M., Duskova, J., Skalova, T., Chon, U., Murakami, K.S., Dohnalek, J., Krasny, L.(2020) Nat Commun 11: 6419-6419
- PubMed: 33339823
- DOI: https://doi.org/10.1038/s41467-020-20158-4
- Primary Citation of Related Structures:
6VSX, 6YXU, 6YYS - PubMed Abstract:
RNA synthesis is central to life, and RNA polymerase (RNAP) depends on accessory factors for recovery from stalled states and adaptation to environmental changes. Here, we investigated the mechanism by which a helicase-like factor HelD recycles RNAP. We report a cryo-EM structure of a complex between the Mycobacterium smegmatis RNAP and HelD. The crescent-shaped HelD simultaneously penetrates deep into two RNAP channels that are responsible for nucleic acids binding and substrate delivery to the active site, thereby locking RNAP in an inactive state. We show that HelD prevents non-specific interactions between RNAP and DNA and dissociates stalled transcription elongation complexes. The liberated RNAP can either stay dormant, sequestered by HelD, or upon HelD release, restart transcription. Our results provide insights into the architecture and regulation of the highly medically-relevant mycobacterial transcription machinery and define HelD as a clearing factor that releases RNAP from nonfunctional complexes with nucleic acids.
- EMBL Grenoble, 71 Avenue des Martyrs, Grenoble, France. tkouba@embl.fr.
Organizational Affiliation: