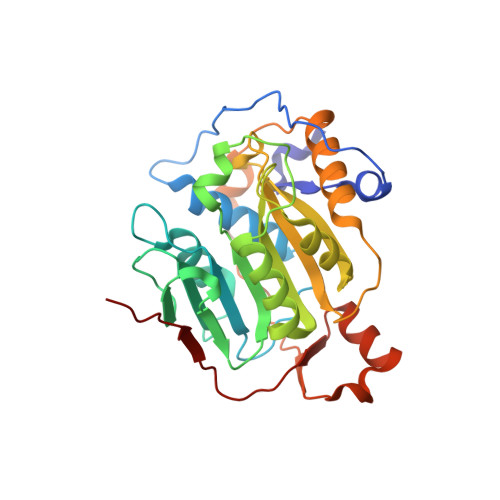
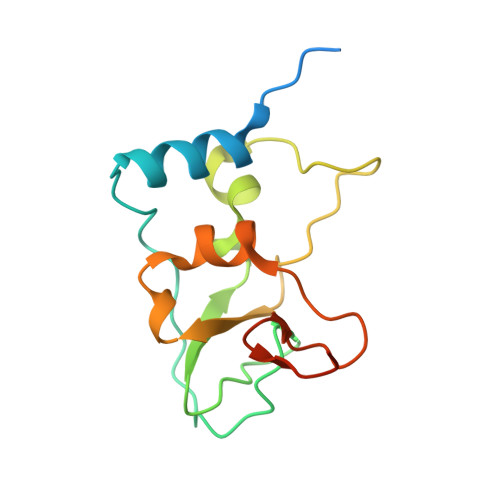
Structural basis of RNA cap modification by SARS-CoV-2.
Viswanathan, T., Arya, S., Chan, S.H., Qi, S., Dai, N., Misra, A., Park, J.G., Oladunni, F., Kovalskyy, D., Hromas, R.A., Martinez-Sobrido, L., Gupta, Y.K.(2020) Nat Commun 11: 3718-3718
- PubMed: 32709886
- DOI: https://doi.org/10.1038/s41467-020-17496-8
- Primary Citation of Related Structures:
6WKS - PubMed Abstract:
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causative agent of COVID-19 illness, has caused millions of infections worldwide. In SARS coronaviruses, the non-structural protein 16 (nsp16), in conjunction with nsp10, methylates the 5'-end of virally encoded mRNAs to mimic cellular mRNAs, thus protecting the virus from host innate immune restriction. We report here the high-resolution structure of a ternary complex of SARS-CoV-2 nsp16 and nsp10 in the presence of cognate RNA substrate analogue and methyl donor, S-adenosyl methionine (SAM). The nsp16/nsp10 heterodimer is captured in the act of 2'-O methylation of the ribose sugar of the first nucleotide of SARS-CoV-2 mRNA. We observe large conformational changes associated with substrate binding as the enzyme transitions from a binary to a ternary state. This induced fit model provides mechanistic insights into the 2'-O methylation of the viral mRNA cap. We also discover a distant (25 Å) ligand-binding site unique to SARS-CoV-2, which can alternatively be targeted, in addition to RNA cap and SAM pockets, for antiviral development.
- Greehey Children's Cancer Research Institute, University of Texas Health at San Antonio, 8403 Floyd Curl Drive, San Antonio, TX, 78229, USA.
Organizational Affiliation: