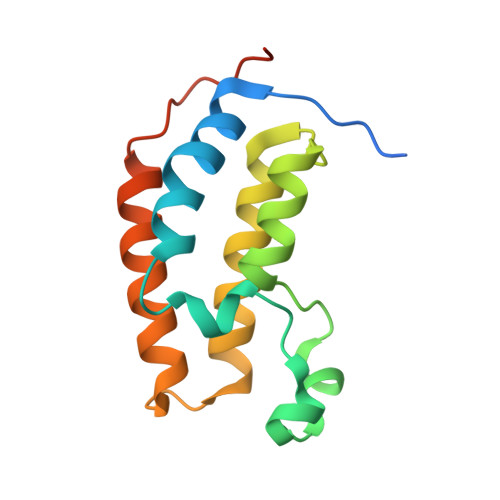
Cyclic peptides can engage a single binding pocket through highly divergent modes.
Patel, K., Walport, L.J., Walshe, J.L., Solomon, P.D., Low, J.K.K., Tran, D.H., Mouradian, K.S., Silva, A.P.G., Wilkinson-White, L., Norman, A., Franck, C., Matthews, J.M., Guss, J.M., Payne, R.J., Passioura, T., Suga, H., Mackay, J.P.(2020) Proc Natl Acad Sci U S A 117: 26728-26738
- PubMed: 33046654
- DOI: https://doi.org/10.1073/pnas.2003086117
- Primary Citation of Related Structures:
6U4A, 6U61, 6U6K, 6U6L, 6U71, 6U72, 6U74, 6U8G, 6U8H, 6U8I, 6U8M, 6ULP, 6ULQ, 6ULT, 6ULV - PubMed Abstract:
Cyclic peptide library screening technologies show immense promise for identifying drug leads and chemical probes for challenging targets. However, the structural and functional diversity encoded within such libraries is largely undefined. We have systematically profiled the affinity, selectivity, and structural features of library-derived cyclic peptides selected to recognize three closely related targets: the acetyllysine-binding bromodomain proteins BRD2, -3, and -4. We report affinities as low as 100 pM and specificities of up to 10 6 -fold. Crystal structures of 13 peptide-bromodomain complexes reveal remarkable diversity in both structure and binding mode, including both α-helical and β-sheet structures as well as bivalent binding modes. The peptides can also exhibit a high degree of structural preorganization. Our data demonstrate the enormous potential within these libraries to provide diverse binding modes against a single target, which underpins their capacity to yield highly potent and selective ligands.
Organizational Affiliation:
School of Life and Environmental Sciences, The University of Sydney, Sydney, NSW 2006, Australia.