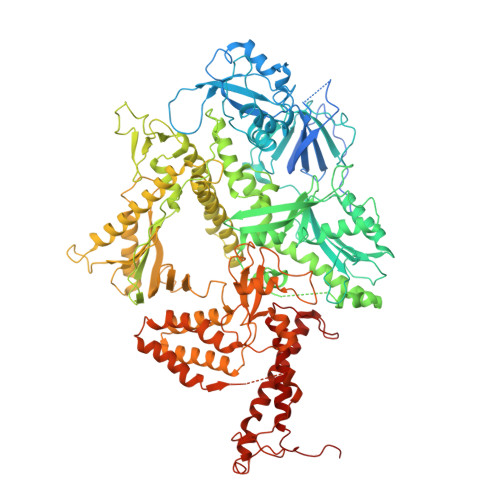
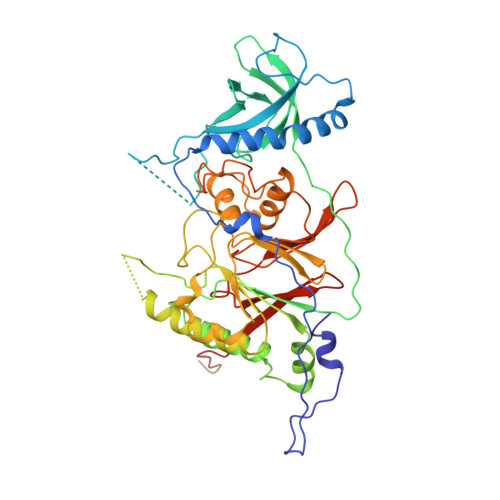
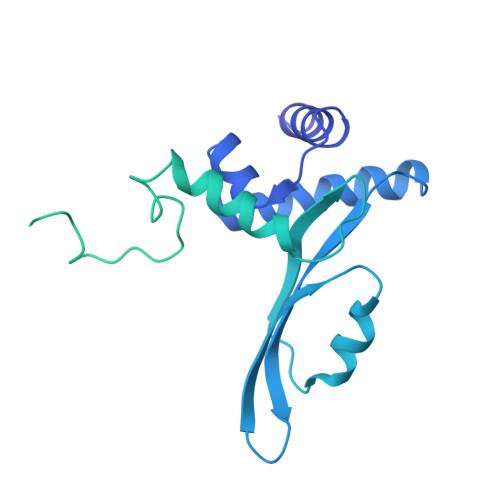
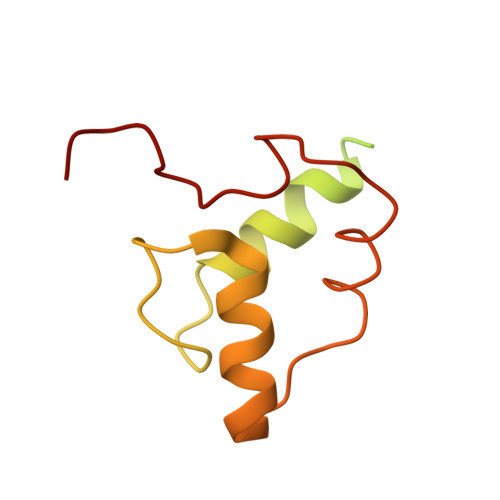
Structure of the processive human Pol delta holoenzyme.
Lancey, C., Tehseen, M., Raducanu, V.S., Rashid, F., Merino, N., Ragan, T.J., Savva, C.G., Zaher, M.S., Shirbini, A., Blanco, F.J., Hamdan, S.M., De Biasio, A.(2020) Nat Commun 11: 1109-1109
- PubMed: 32111820
- DOI: https://doi.org/10.1038/s41467-020-14898-6
- Primary Citation of Related Structures:
6S1M, 6S1N, 6S1O, 6TNY, 6TNZ - PubMed Abstract:
In eukaryotes, DNA polymerase δ (Pol δ) bound to the proliferating cell nuclear antigen (PCNA) replicates the lagging strand and cooperates with flap endonuclease 1 (FEN1) to process the Okazaki fragments for their ligation. We present the high-resolution cryo-EM structure of the human processive Pol δ-DNA-PCNA complex in the absence and presence of FEN1. Pol δ is anchored to one of the three PCNA monomers through the C-terminal domain of the catalytic subunit. The catalytic core sits on top of PCNA in an open configuration while the regulatory subunits project laterally. This arrangement allows PCNA to thread and stabilize the DNA exiting the catalytic cleft and recruit FEN1 to one unoccupied monomer in a toolbelt fashion. Alternative holoenzyme conformations reveal important functional interactions that maintain PCNA orientation during synthesis. This work sheds light on the structural basis of Pol δ's activity in replicating the human genome.
- Leicester Institute of Structural & Chemical Biology and Department of Molecular & Cell Biology, University of Leicester, Lancaster Rd, Leicester, LE1 7HB, UK.
Organizational Affiliation: