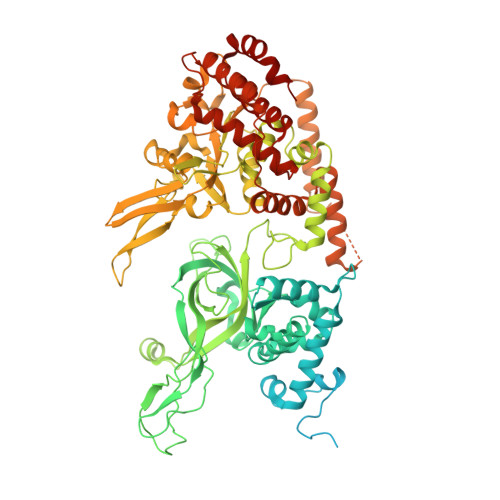
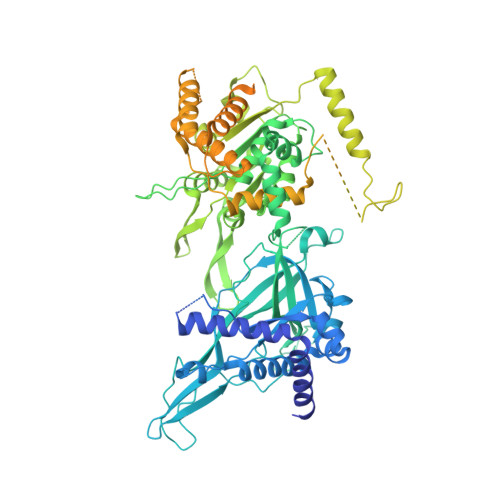
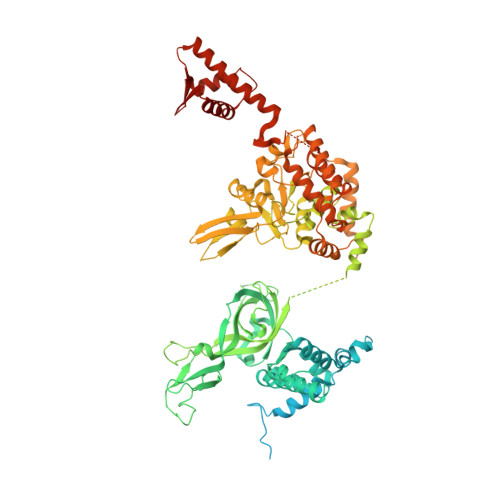
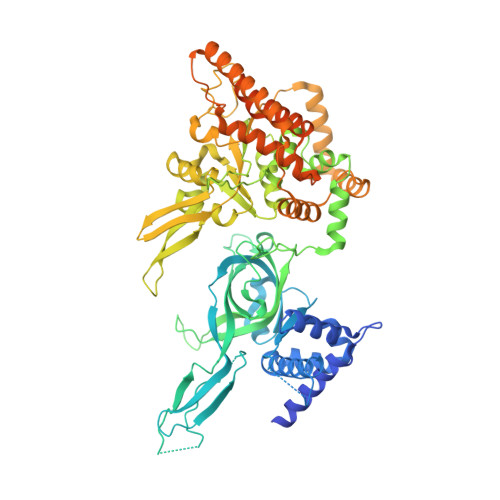
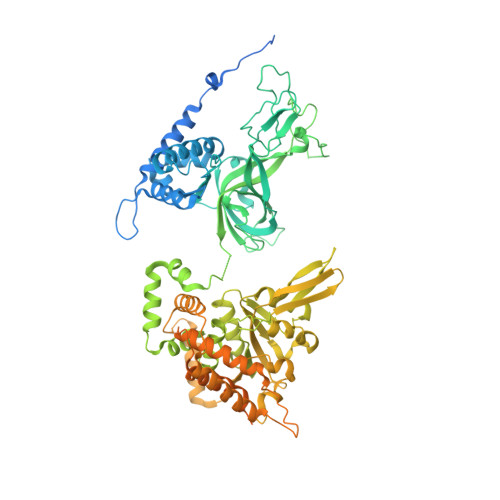
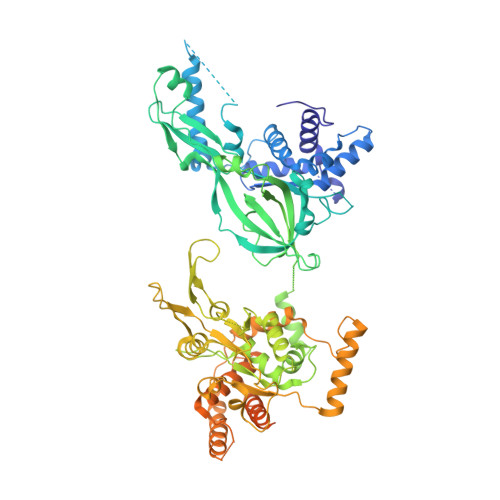
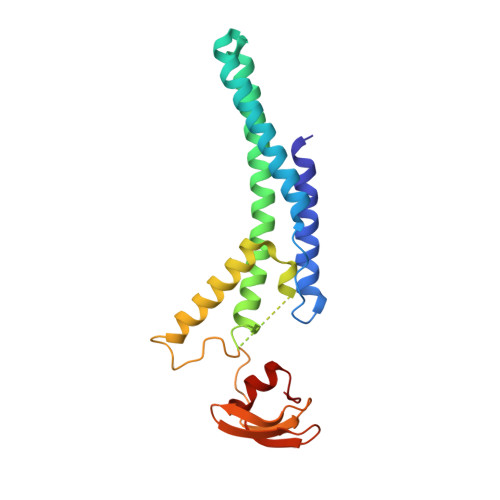
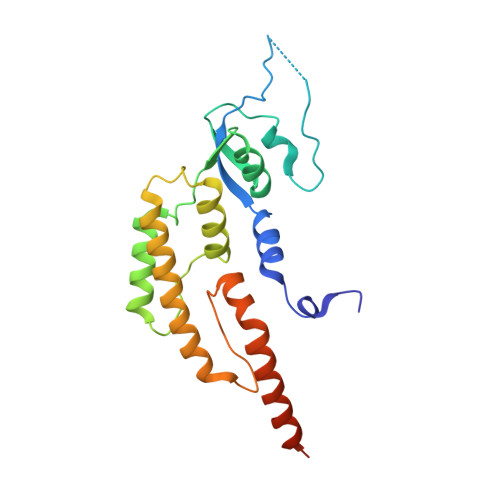
Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Baretic, D., Jenkyn-Bedford, M., Aria, V., Cannone, G., Skehel, M., Yeeles, J.T.P.(2020) Mol Cell 78: 926-940.e13
- PubMed: 32369734
- DOI: https://doi.org/10.1016/j.molcel.2020.04.012
- Primary Citation of Related Structures:
6SKL, 6SKO - PubMed Abstract:
The eukaryotic replisome, organized around the Cdc45-MCM-GINS (CMG) helicase, orchestrates chromosome replication. Multiple factors associate directly with CMG, including Ctf4 and the heterotrimeric fork protection complex (Csm3/Tof1 and Mrc1), which has important roles including aiding normal replication rates and stabilizing stalled forks. How these proteins interface with CMG to execute these functions is poorly understood. Here we present 3 to 3.5 Å resolution electron cryomicroscopy (cryo-EM) structures comprising CMG, Ctf4, and the fork protection complex at a replication fork. The structures provide high-resolution views of CMG-DNA interactions, revealing a mechanism for strand separation, and show Csm3/Tof1 "grip" duplex DNA ahead of CMG via a network of interactions important for efficient replication fork pausing. Although Mrc1 was not resolved in our structures, we determine its topology in the replisome by cross-linking mass spectrometry. Collectively, our work reveals how four highly conserved replisome components collaborate with CMG to facilitate replisome progression and maintain genome stability.
- Laboratory of Molecular Biology, Medical Research Council, Cambridge CB2 0QH, UK.
Organizational Affiliation: