Structural characterization of kiteplatinated DNA
Krejcikova, M., Papadia, P., Gkionis, K., Plats, J., Petruzzella, E., Savino, S., Hoeschelle, J.D., Natile, G., Sponer, J., Kubicek, K., Margiotta, N.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| Kiteplatinated DNA oligomer, chain A | 12 | Homo sapiens | 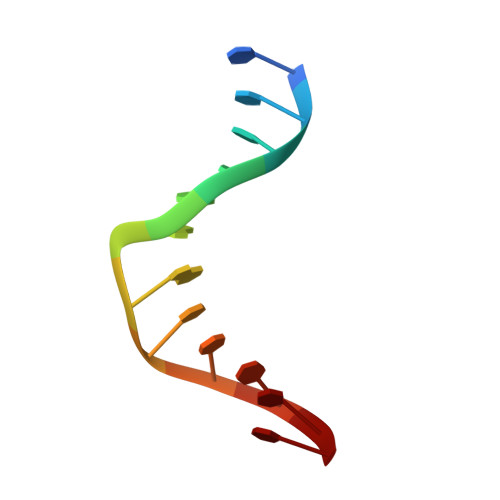 | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| Kiteplatinated DNA oligomer, chain B | 12 | Homo sapiens | 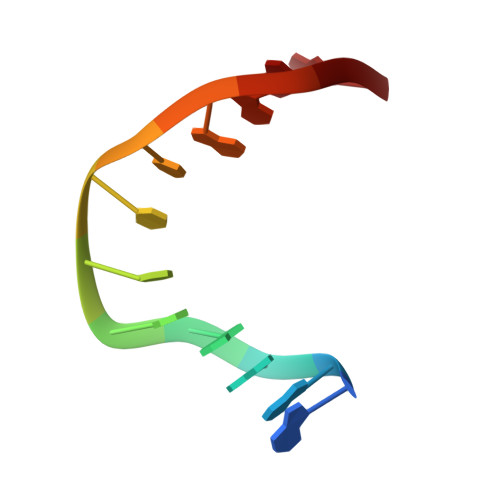 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LN8 (Subject of Investigation/LOI) Query on LN8 | C [auth A] | Kiteplatin C6 H14 Cl2 N2 Pt CPGREGRXVKABHL-UOXVPMNTSA-L |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Education (Czech Republic) | Czech Republic | LQ1601 |