Biophysical Consequences of EVEN-PLUS Syndrome Mutations for the Function of Mortalin.
Moseng, M.A., Nix, J.C., Page, R.C.(2019) J Phys Chem B 123: 3383-3396
- PubMed: 30933555
- DOI: https://doi.org/10.1021/acs.jpcb.9b00071
- Primary Citation of Related Structures:
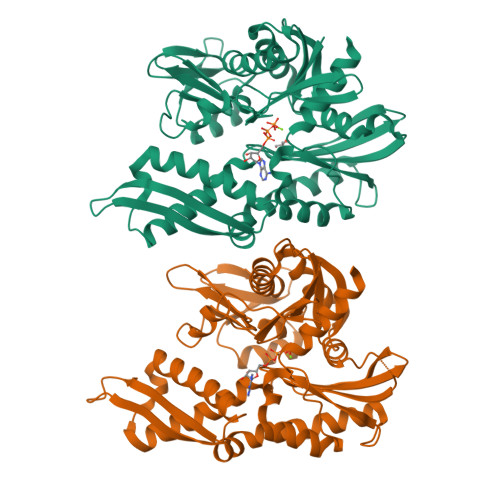
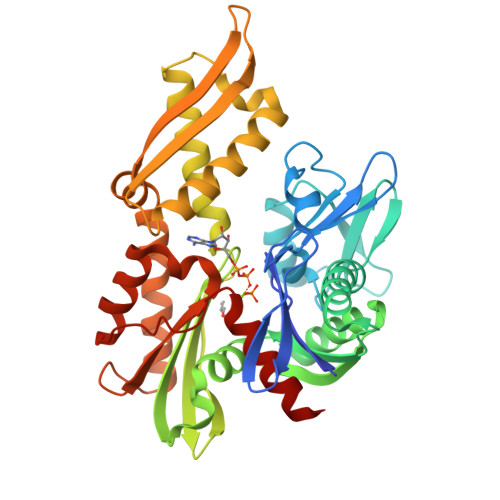
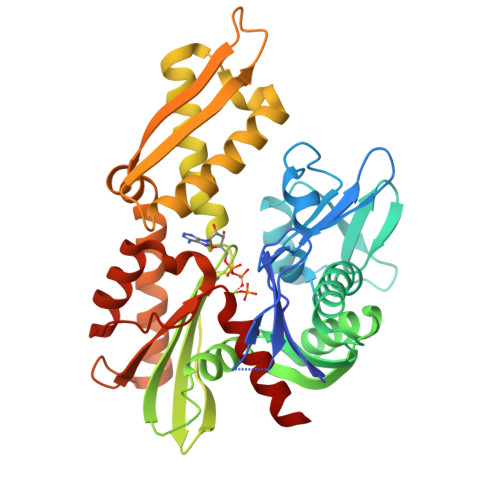
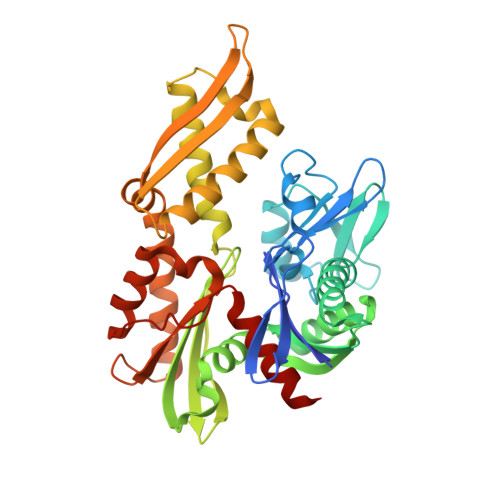
6NHK - PubMed Abstract:
HSPA9, the gene coding for the mitochondrial chaperone mortalin, is involved in various cellular roles such as mitochondrial protein import, folding, degradation, Fe-S cluster biogenesis, mitochondrial homeostasis, and regulation of the antiapoptotic protein p53. Mutations in the HSPA9 gene, particularly within the region coding for the nucleotide-binding domain (NBD), cause the autosomal disorder known as EVEN-PLUS syndrome. The resulting mutants R126W and Y128C are located on the surface of the mortalin-NBD near the binding interface with the interdomain linker (IDL). We used differential scanning fluorimetry (DSF), biolayer interferometry, X-ray crystallography, ATP hydrolysis assays, and Rosetta docking simulations to study the structural and functional consequences of the EVEN-PLUS syndrome-associated R126W and Y128C mutations within the mortalin-NBD. These results indicate that the surface mutations R126W and Y128C have far-reaching effects that disrupt ATP hydrolysis, interdomain linker binding, and thermostability and increase the propensity for aggregation. The structural differences observed provide insight into how the conformations of mortalin differ from other heat shock protein 70 (Hsp70) homologues. Combined, our biophysical and structural studies contribute to the understanding of the molecular basis for how disease-associated mortalin mutations affect mortalin functionality and the pathogenesis of EVEN-PLUS syndrome.
Organizational Affiliation:
Department of Chemistry and Biochemistry , Miami University , Oxford , Ohio 45056 , United States.