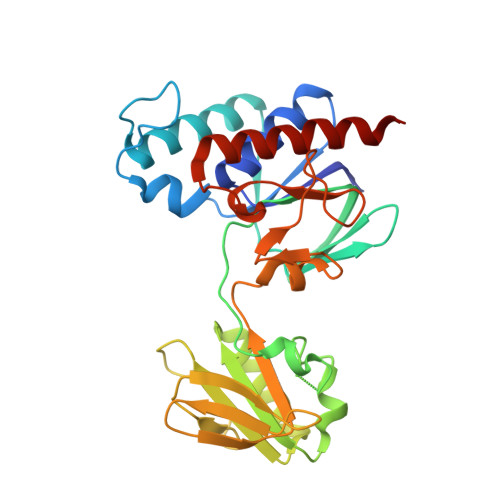
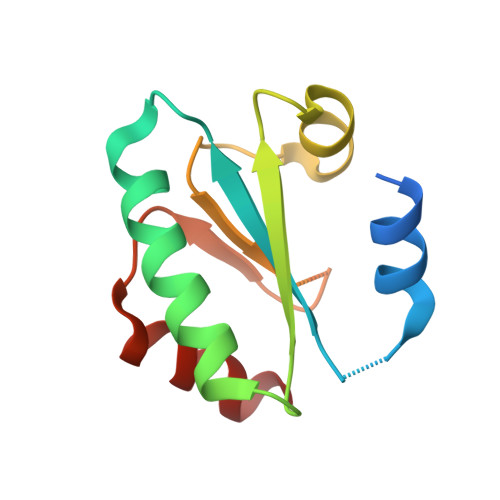
Ferredoxin-linked flavoenzyme defines a family of pyridine nucleotide-independent thioredoxin reductases.
Buey, R.M., Fernandez-Justel, D., de Pereda, J.M., Revuelta, J.L., Schurmann, P., Buchanan, B.B., Balsera, M.(2018) Proc Natl Acad Sci U S A 115: 12967-12972
- PubMed: 30510005
- DOI: https://doi.org/10.1073/pnas.1812781115
- Primary Citation of Related Structures:
6GN9, 6GNA, 6GNB, 6GNC, 6GND - PubMed Abstract:
Ferredoxin-dependent thioredoxin reductase was identified 35 y ago in the fermentative bacterium Clostridium pasteurianum [Hammel KE, Cornwell KL, Buchanan BB (1983) Proc Natl Acad Sci USA 80:3681-3685]. The enzyme, a flavoprotein, was strictly dependent on ferredoxin as reductant and was inactive with either NADPH or NADH. This early work has not been further pursued. We have recently reinvestigated the problem and confirmed that the enzyme, here designated ferredoxin-dependent flavin thioredoxin reductase (FFTR), is a flavoprotein. The enzyme differs from ferredoxin-thioredoxin reductase (FTR), which has a signature [4Fe-4S] cluster, but shows structural similarities to NADP-dependent thioredoxin reductase (NTR). Comparative amino acid sequence analysis showed that FFTR is present in a number of clostridial species, some of which lack both FTR and an archetypal NTR. We have isolated, crystallized, and determined the structural properties of FFTR from a member of this group, Clostridium acetobutylicum , both alone and in complex with Trx. The structures showed an elongated FFTR homodimer, each monomer comprising two Rossmann domains and a noncovalently bound FAD cofactor that exposes the isoalloxazine ring to the solvent. The FFTR structures revealed an alternative domain organization compared with NTR that enables the enzyme to accommodate Fdx rather than NADPH. The results suggest that FFTR exists in a range of conformations with varying degrees of domain separation in solution and that the stacking between the two redox-active groups for the transfer of reducing equivalents results in a profound structural reorganization. A mechanism in accord with the findings is proposed.
- Metabolic Engineering Group, Departamento de Microbiología y Genética, Universidad de Salamanca, 37007 Salamanca, Spain.
Organizational Affiliation: