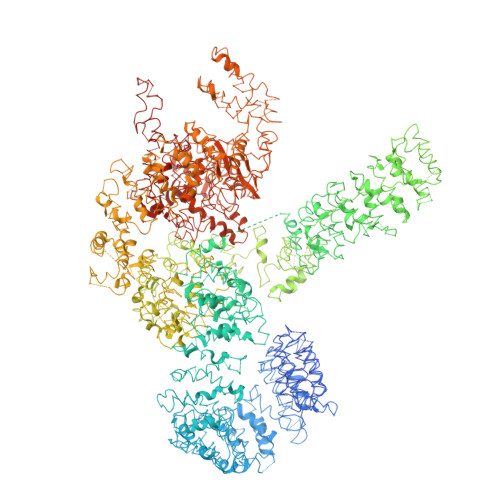
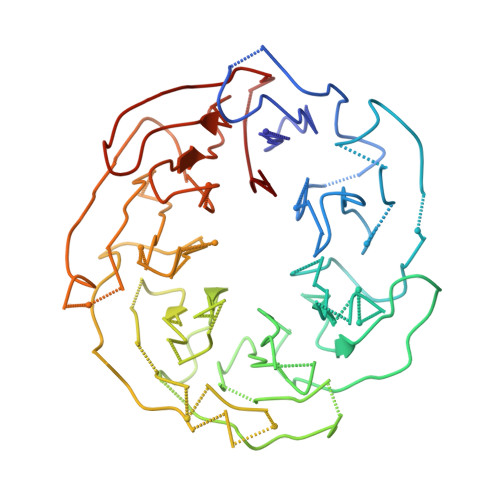
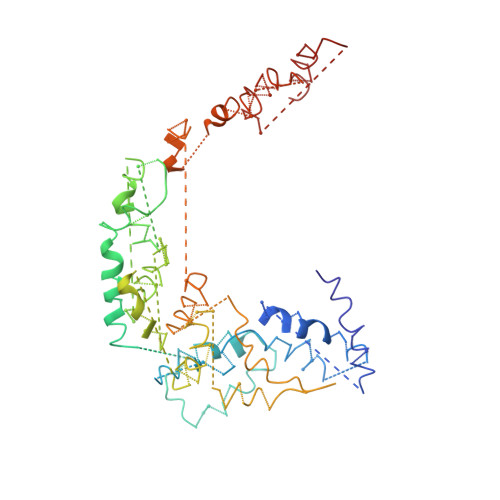
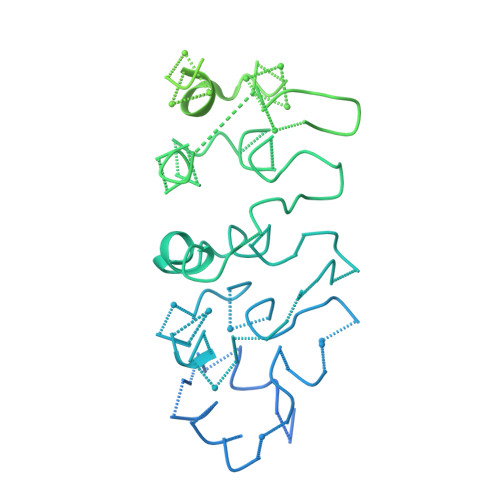
Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2.
Karuppasamy, M., Kusmider, B., Oliveira, T.M., Gaubitz, C., Prouteau, M., Loewith, R., Schaffitzel, C.(2017) Nat Commun 8: 1729-1729
- PubMed: 29170376
- DOI: https://doi.org/10.1038/s41467-017-01862-0
- Primary Citation of Related Structures:
6EMK - PubMed Abstract:
The target of rapamycin (TOR) kinase assembles into two distinct multiprotein complexes, conserved across eukaryote evolution. In contrast to TOR complex 1 (TORC1), TORC2 kinase activity is not inhibited by the macrolide rapamycin. Here, we present the structure of Saccharomyces cerevisiae TORC2 determined by electron cryo-microscopy. TORC2 contains six subunits assembling into a 1.4 MDa rhombohedron. Tor2 and Lst8 form the common core of both TOR complexes. Avo3/Rictor is unique to TORC2, but interacts with the same HEAT repeats of Tor2 that are engaged by Kog1/Raptor in mammalian TORC1, explaining the mutual exclusivity of these two proteins. Density, which we conclude is Avo3, occludes the FKBP12-rapamycin-binding site of Tor2's FRB domain rendering TORC2 rapamycin insensitive and recessing the kinase active site. Although mobile, Avo1/hSin1 further restricts access to the active site as its conserved-region-in-the-middle (CRIM) domain is positioned along an edge of the TORC2 active-site-cleft, consistent with a role for CRIM in substrate recruitment.
- European Molecular Biology Laboratory, Grenoble Outstation, 71 Avenue des Martyrs, 38042, Grenoble, France.
Organizational Affiliation: