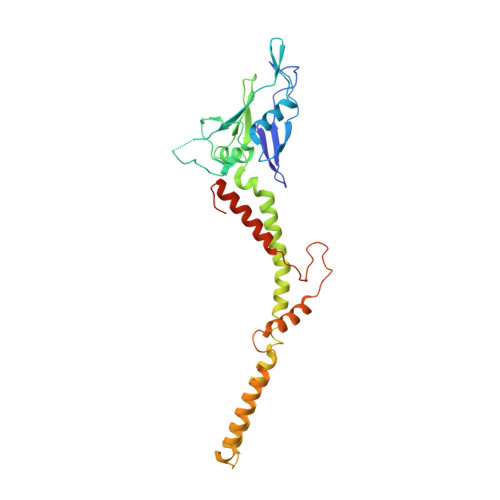
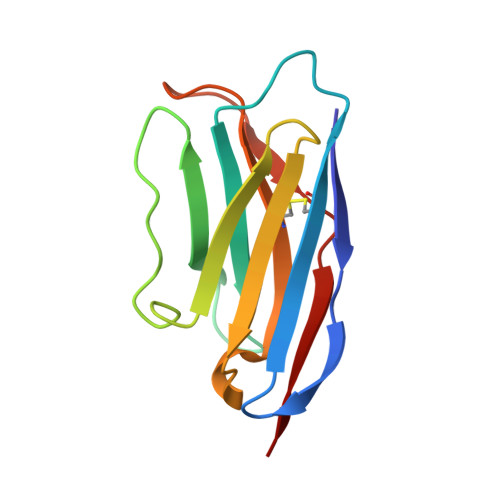
X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Fan, C., Fan, M., Orlando, B.J., Fastman, N.M., Zhang, J., Xu, Y., Chambers, M.G., Xu, X., Perry, K., Liao, M., Feng, L.(2018) Nature 559: 575-579
- PubMed: 29995856
- DOI: https://doi.org/10.1038/s41586-018-0330-9
- Primary Citation of Related Structures:
6C5R, 6C5W - PubMed Abstract:
Mitochondrial calcium uptake is critical for regulating ATP production, intracellular calcium signalling, and cell death. This uptake is mediated by a highly selective calcium channel called the mitochondrial calcium uniporter (MCU). Here, we determined the structures of the pore-forming MCU proteins from two fungi by X-ray crystallography and single-particle cryo-electron microscopy. The stoichiometry, overall architecture, and individual subunit structure differed markedly from those described in the recent nuclear magnetic resonance structure of Caenorhabditis elegans MCU. We observed a dimer-of-dimer architecture across species and chemical environments, which was corroborated by biochemical experiments. Structural analyses and functional characterization uncovered the roles of key residues in the pore. These results reveal a new ion channel architecture, provide insights into calcium coordination, selectivity and conduction, and establish a structural framework for understanding the mechanism of mitochondrial calcium uniporter function.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, USA.
Organizational Affiliation: