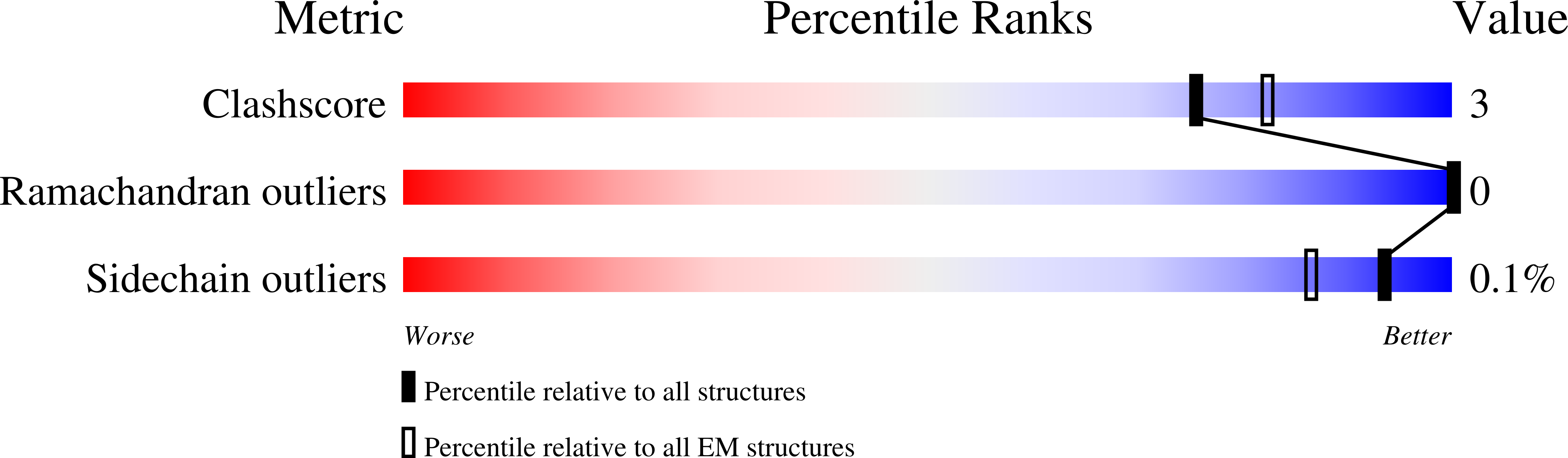ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component G [auth B] ,H [auth C] 105 Escherichia coli Mutation(s) : 0 Gene Names: mlaB ,
yrbB ,
A6581_13415 ,
A6592_14225 ,
A6V01_00625 ,
A8C65_02170 ,
A9P13_02945 ,
A9R57_12600 ,
A9X72_02835 ,
AC067_23675 ,
ACN002_3282 ,
ACN68_12355 ,
ACN81_18445 ,
ACU57_14445 ,
ACU90_11565 ,
AM270_15510 ,
AM446_04320 ,
AM464_08585 ,
AMK83_06340 ,
AML07_19340 ,
APT94_23200 ,
APZ14_04295 ,
ARC77_12405 ,
AU473_05735 ,
AUQ13_03075 ,
AUS26_18260 ,
AW106_13275 ,
AWB10_02850 ,
AWG90_020620 ,
AWP75_14100 ,
B9M99_10250 ,
B9T59_16250 ,
BANRA_01811 ,
BANRA_03804 ,
BANRA_03999 ,
BANRA_04207 ,
BB545_09780 ,
BE963_06830 ,
BER14_09135 ,
BHJ80_21985 ,
BHS81_19200 ,
BHS87_17970 ,
BIQ87_18040 ,
BIU72_05405 ,
BJJ90_02770 ,
BK292_10220 ,
BK296_13050 ,
BK373_10175 ,
BK375_18740 ,
BK383_15965 ,
BMA87_01295 ,
BMT49_25595 ,
BMT91_11610 ,
BOH76_04915 ,
BON63_14235 ,
BON66_12205 ,
BON69_26555 ,
BON71_25170 ,
BON72_13935 ,
BON75_23145 ,
BON76_05525 ,
BON83_09240 ,
BON86_01565 ,
BON87_18195 ,
BON92_11035 ,
BON94_12335 ,
BON95_22020 ,
BON96_06355 ,
BTQ04_16475 ,
BTQ06_11870 ,
BUE81_19690 ,
BvCms2454_04822 ,
BvCms28BK_03527 ,
BvCms35BK_01543 ,
BvCmsHHP001_01202 ,
BvCmsHHP019_05314 ,
BvCmsKKP061_00888 ,
BvCmsKSNP073_02163 ,
BvCmsKSNP081_00287 ,
BvCmsKSP011_02050 ,
BvCmsKSP024_00283 ,
BvCmsKSP026_00502 ,
BvCmsKSP045_01650 ,
BvCmsKSP067_01464 ,
BvCmsNSNP036_02151 ,
BvCmsNSP006_04640 ,
BvCmsNSP047_04118 ,
BvCmsNSP072_03187 ,
BvCmsOUP014_03632 ,
BvCmsSINP011_04458 ,
BvCmsSIP019_00407 ,
BvCmsSIP044_00137 ,
BVL39_10130 ,
BW690_19360 ,
BWI89_22070 ,
BWP17_10155 ,
BXT93_02550 ,
BZL31_05925 ,
BZL69_07715 ,
C2M16_04215 ,
C5715_22795 ,
C5N07_18070 ,
C5P01_18835 ,
C6669_12020 ,
C7235_03225 ,
C7B02_15875 ,
C7B08_03565 ,
C7B18_13830 ,
C9098_07790 ,
C9114_15455 ,
C9141_00390 ,
C9160_02340 ,
C9162_04110 ,
C9201_13875 ,
C9306_00415 ,
C9E25_11270 ,
C9E67_03375 ,
C9Z03_07490 ,
C9Z28_00385 ,
C9Z29_07190 ,
C9Z37_14660 ,
C9Z39_14275 ,
C9Z43_00380 ,
C9Z68_06245 ,
C9Z69_11175 ,
C9Z70_06765 ,
C9Z78_03315 ,
C9Z89_09065 ,
CA593_10345 ,
CDL37_12215 ,
CF006_16665 ,
CG692_07300 ,
CI641_009255 ,
CI693_03675 ,
CI694_20835 ,
CIG45_16045 ,
CJU63_19140 ,
CJU64_19020 ,
CMR93_08835 ,
CO706_14630 ,
COD30_12915 ,
COD46_10760 ,
CQP61_03650 ,
CR538_03070 ,
CR539_21250 ,
CRD98_04715 ,
CRE06_06830 ,
CRM83_23500 ,
CRX46_03745 ,
CVH05_10780 ,
CWM24_22870 ,
CWS33_08375 ,
D0X26_09090 ,
D2184_10505 ,
D2185_00445 ,
D2188_02580 ,
D3821_08325 ,
D3822_14575 ,
D3O91_07625 ,
D3P01_09160 ,
D3Y67_18290 ,
D4011_13725 ,
D4074_02435 ,
D4628_02655 ,
D4636_02890 ,
D4638_00790 ,
D4660_02830 ,
D4718_09085 ,
D4L91_11950 ,
D4M06_11225 ,
D4U85_14330 ,
D5H35_09780 ,
D5I97_10670 ,
D6004_08125 ,
D6C36_14160 ,
D6D43_15510 ,
D6T60_15390 ,
D6T98_02410 ,
D6W00_14015 ,
D6X36_05360 ,
D6X63_01430 ,
D6X76_04335 ,
D7K33_12855 ,
D7K63_12220 ,
D7K66_04495 ,
D7W70_15855 ,
D7Z75_02945 ,
D8Y65_13475 ,
D9610_07810 ,
D9C99_05120 ,
D9D20_06275 ,
D9D31_14515 ,
D9D43_03615 ,
D9D44_04410 ,
D9E19_10400 ,
D9E34_11740 ,
D9E49_09595 ,
D9F17_07200 ,
D9F32_01240 ,
D9F87_05870 ,
D9G29_07025 ,
D9G48_11290 ,
D9G69_01100 ,
D9G95_17085 ,
D9H53_01195 ,
D9H68_07560 ,
D9I18_07175 ,
D9I88_07240 ,
D9J03_05150 ,
D9J11_01130 ,
D9J44_07850 ,
D9J52_07915 ,
D9J58_06110 ,
D9J60_11185 ,
D9J63_16910 ,
D9J78_05130 ,
D9K02_06080 ,
D9K48_12255 ,
D9L89_05370 ,
D9S45_00255 ,
D9X77_02330 ,
D9X97_03120 ,
D9Z28_03370 ,
DAH18_20610 ,
DAH26_17315 ,
DAH30_12875 ,
DAH32_13975 ,
DAH34_01980 ,
DAH37_04255 ,
DAH43_16780 ,
DBQ99_04040 ,
DD762_13125 ,
DEN86_08060 ,
DEN89_09405 ,
DEN97_16135 ,
DEO04_19605 ,
DEO19_06430 ,
DIV22_02975 ,
DJ503_19040 ,
DK132_07375 ,
DL257_05650 ,
DL292_05795 ,
DL326_06060 ,
DL455_12700 ,
DL479_06590 ,
DL530_10055 ,
DL705_01300 ,
DL800_23275 ,
DLT82_02655 ,
DLU50_08550 ,
DLU67_08375 ,
DLU82_10330 ,
DLW60_07490 ,
DLW88_04795 ,
DLY41_08155 ,
DM102_15165 ,
DM155_07645 ,
DM267_03265 ,
DM280_09055 ,
DM296_04835 ,
DM382_07360 ,
DM820_04625 ,
DM962_02640 ,
DM973_04650 ,
DMI04_04270 ,
DMI53_12715 ,
DMO02_15405 ,
DMY83_09920 ,
DN660_01330 ,
DN700_04595 ,
DN703_01080 ,
DN808_01890 ,
DNB37_13705 ,
DNC98_07160 ,
DND16_06535 ,
DND79_01330 ,
DNI21_03635 ,
DNJ62_12070 ,
DNK12_02835 ,
DNQ45_01620 ,
DNW42_05405 ,
DNX19_05580 ,
DNX30_11365 ,
DOE35_12610 ,
DOS18_13030 ,
DOT81_04255 ,
DOU81_12375 ,
DOY22_14085 ,
DOY56_12245 ,
DOY61_04695 ,
DOY67_01285 ,
DP265_06555 ,
DP277_10105 ,
DQE83_16545 ,
DQE91_01265 ,
DQF36_08635 ,
DQF57_03615 ,
DQF71_07160 ,
DQF72_06460 ,
DQG35_02830 ,
DQO13_01295 ,
DQP61_01770 ,
DRP48_13910 ,
DRW19_01295 ,
DS143_06770 ,
DS721_11090 ,
DS732_23695 ,
DT034_01270 ,
DTL43_04295 ,
DTM10_06985 ,
DTM45_14510 ,
DTZ20_12090 ,
DU309_11135 ,
DU321_08670 ,
DVB38_07395 ,
DXT69_05520 ,
DXT71_00440 ,
DXX80_009590 ,
E0I42_14340 ,
E0K84_17700 ,
E0L12_03915 ,
E2119_05870 ,
E2127_02575 ,
E2128_02495 ,
E2129_05760 ,
E2134_15595 ,
E2135_08080 ,
E2855_04145 ,
E2863_03970 ,
E4K55_06660 ,
E4K61_09280 ,
E5P22_07075 ,
E5P28_12910 ,
E5P37_13135 ,
E5S42_17100 ,
E5S46_15725 ,
E5S47_16890 ,
E5S58_02935 ,
E5S61_19420 ,
EA214_08600 ,
EA223_04825 ,
EA225_07080 ,
EA233_06175 ,
EAI42_05495 ,
EAI52_09890 ,
EAM59_01020 ,
EAN70_10745 ,
EAX79_13230 ,
EB476_09250 ,
EB509_07200 ,
EB510_12405 ,
EB515_07025 ,
EBA84_05830 ,
EBJ06_06135 ,
EBM08_05455 ,
EC1094V2_454 ,
EC3234A_53c00700 ,
EC3426_04342 ,
EC95NR1_02575 ,
ECTO6_00563 ,
ED225_04270 ,
ED307_11785 ,
ED600_09400 ,
ED607_10425 ,
ED611_04280 ,
ED903_04655 ,
ED944_07260 ,
EEA45_06940 ,
EEP23_06300 ,
EF082_03990 ,
EF173_17005 ,
EG075_10110 ,
EG599_06165 ,
EG796_01190 ,
EG808_04845 ,
EGC26_04915 ,
EGU87_04215 ,
EH186_08760 ,
EH412_00995 ,
EHD45_17860 ,
EHD63_05275 ,
EHD79_13055 ,
EHJ36_00995 ,
EHJ66_06090 ,
EHV81_00555 ,
EHV90_06170 ,
EHW09_05600 ,
EHX09_11795 ,
EI021_04065 ,
EI028_15050 ,
EI032_12380 ,
EI041_02000 ,
EIZ86_06785 ,
EIZ93_11790 ,
EJ366_25995 ,
EJC75_15225 ,
EKI52_18235 ,
EL75_0496 ,
EL79_0518 ,
EL80_0509 ,
ELT20_05045 ,
ELU85_06925 ,
ELV05_02275 ,
ELV08_02985 ,
ELV15_01830 ,
ELV28_06345 ,
EPT01_00415 ,
EQ820_11545 ,
EQ823_05050 ,
EQ830_04520 ,
ERL57_13435 ,
ERS085365_00538 ,
ERS085366_00551 ,
ERS085374_01930 ,
ERS085379_00371 ,
ERS085383_00695 ,
ERS085404_02185 ,
ERS085406_02933 ,
ERS085416_00356 ,
ERS139211_02331 ,
ERS150873_02331 ,
ERS150876_01029 ,
EST51_02745 ,
EVY14_07725 ,
EXM29_19995 ,
ExPECSC019_00712 ,
ExPECSC038_02702 ,
EXX71_13400 ,
EXX78_16730 ,
EYD11_02645 ,
EYX82_02455 ,
EYY34_10615 ,
F1E19_20375 ,
F7F00_12170 ,
F7F11_09035 ,
F7F18_01205 ,
F7F23_11445 ,
F7F26_00405 ,
F7F29_05115 ,
F7G01_13980 ,
F7G03_16150 ,
F9Z74_11630 ,
FAF34_018885 ,
FE846_13735 ,
FKO60_06240 ,
FNJ69_14865 ,
FNJ83_20565 ,
FORC82_0569 ,
FQ022_08190 ,
FQ915_12490 ,
FQR64_03035 ,
FQZ46_16130 ,
FTV92_21230 ,
FV293_08375 ,
FVB16_04440 ,
FWK02_19645 ,
FY127_07380 ,
FZ043_22035 ,
GHR40_07595 ,
GII67_12750 ,
GIY13_09305 ,
GIY19_08335 ,
GJ11_20810 ,
GJD97_02885 ,
GKE15_04665 ,
GKE22_12205 ,
GKE24_04655 ,
GKE26_04130 ,
GKE29_02675 ,
GKE31_04620 ,
GKE39_10535 ,
GKE46_11210 ,
GKE58_04575 ,
GKE60_04100 ,
GKE64_10535 ,
GKE77_05005 ,
GKE87_00910 ,
GKE93_06185 ,
GKF00_03235 ,
GKF03_05520 ,
GKF28_12045 ,
GKF34_04640 ,
GKF47_04635 ,
GKF74_09220 ,
GKF86_04260 ,
GKF89_14375 ,
GKG12_08325 ,
GN312_06640 ,
GNZ00_05080 ,
GNZ02_01290 ,
GNZ03_06110 ,
GP654_17710 ,
GP661_13045 ,
GP664_12080 ,
GP666_12960 ,
GP689_13875 ,
GP712_11385 ,
GP935_13340 ,
GP946_08670 ,
GP950_18225 ,
GQE30_02570 ,
GQE34_13735 ,
GQE51_04670 ,
GQE64_09725 ,
GQE68_06200 ,
GQE88_20265 ,
GQE93_14850 ,
GQM06_05785 ,
GQM09_11065 ,
GQM13_09420 ,
GQM17_12885 ,
GQN16_09500 ,
GRW42_15935 ,
GRW80_12045 ,
HmCmsJML074_04878 ,
HmCmsJML079_04444 ,
HmCmsJML204_02250 ,
HMPREF3040_00351 ,
HW43_20915 ,
MJ49_14345 ,
NCTC10082_02946 ,
NCTC10090_03637 ,
NCTC10418_00837 ,
NCTC10764_03982 ,
NCTC10766_02903 ,
NCTC10865_00786 ,
NCTC10963_00565 ,
NCTC10974_00682 ,
NCTC11022_03358 ,
NCTC11341_01768 ,
NCTC12650_00838 ,
NCTC12950_00597 ,
NCTC13127_00906 ,
NCTC13216_01244 ,
NCTC7922_03906 ,
NCTC7927_00668 ,
NCTC8179_06070 ,
NCTC8500_00486 ,
NCTC8622_00353 ,
NCTC8960_03188 ,
NCTC8985_05122 ,
NCTC9007_04416 ,
NCTC9044_01237 ,
NCTC9050_03718 ,
NCTC9055_02489 ,
NCTC9062_02576 ,
NCTC9077_00732 ,
NCTC9111_00974 ,
NCTC9117_00877 ,
NCTC9701_00685 ,
NCTC9702_00649 ,
NCTC9703_05076 ,
NCTC9706_02844 ,
NCTC9777_02009 ,
PGD_03924 ,
RG28_21075 ,
RK56_014610 ,
RX35_04279 ,
SAMEA3472043_01825 ,
SAMEA3472047_01897 ,
SAMEA3472055_00376 ,
SAMEA3472070_01148 ,
SAMEA3472080_00969 ,
SAMEA3472090_00572 ,
SAMEA3472110_01519 ,
SAMEA3472112_01503 ,
SAMEA3472114_00459 ,
SAMEA3472147_01902 ,
SAMEA3484427_00584 ,
SAMEA3484429_00758 ,
SAMEA3484434_03617 ,
SAMEA3752372_01803 ,
SAMEA3752553_01163 ,
SAMEA3752557_01851 ,
SAMEA3752559_03249 ,
SAMEA3752620_02365 ,
SAMEA3753064_02960 ,
SAMEA3753164_02324 ,
SAMEA3753290_00124 ,
SAMEA3753300_02107 ,
SK85_03504 ,
UC41_09935 ,
UN86_15380 ,
WQ89_06640 ,
WR15_24440 ,
YDC107_1864