Design, Synthesis, and Pharmacological Characterization of a Neutral, Non-Prodrug Thrombin Inhibitor with Good Oral Pharmacokinetics.
Hillisch, A., Gericke, K.M., Allerheiligen, S., Roehrig, S., Schaefer, M., Tersteegen, A., Schulz, S., Lienau, P., Gnoth, M., Puetter, V., Hillig, R.C., Heitmeier, S.(2020) J Med Chem 63: 12574-12594
- PubMed: 33108181
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01035
- Primary Citation of Related Structures:
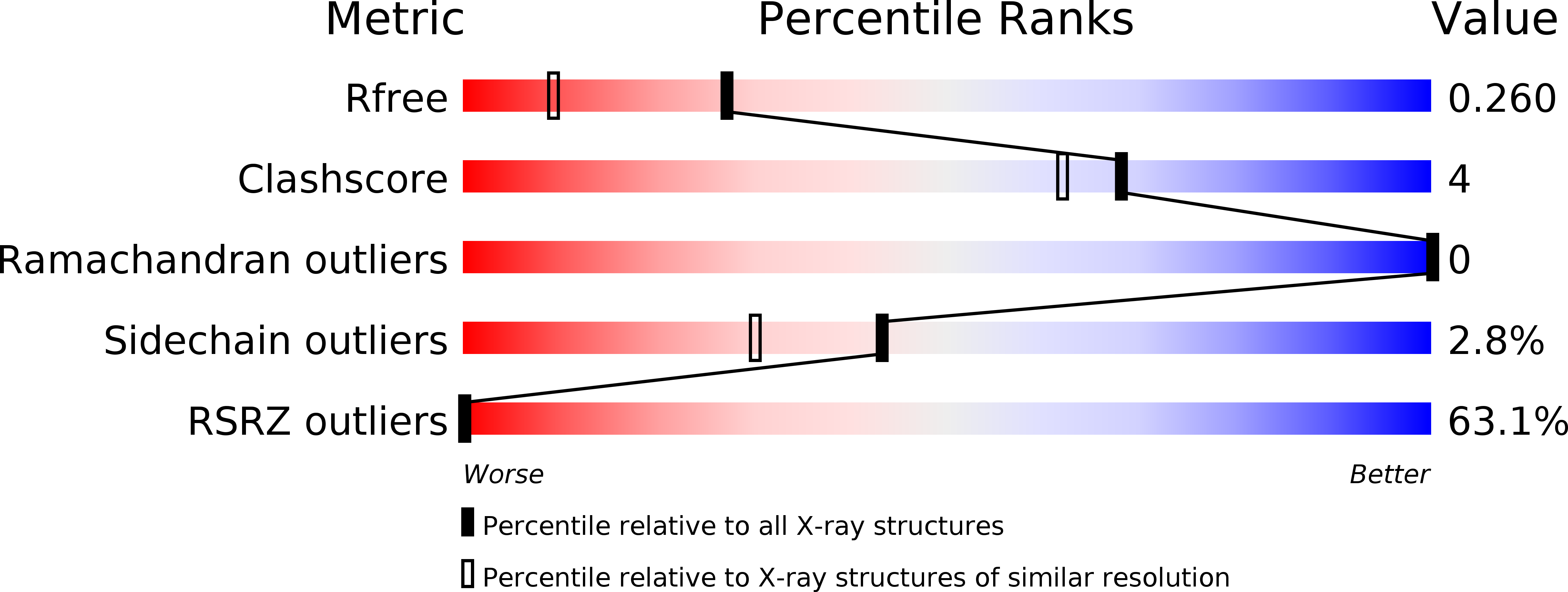
6TFI, 6ZUG, 6ZUH, 6ZUN, 6ZUU, 6ZUW, 6ZUX, 6ZV7, 6ZV8 - PubMed Abstract:
Despite extensive research on small molecule thrombin inhibitors for oral application in the past decades, only a single double prodrug with very modest oral bioavailability has reached human therapy as a marketed drug. We have undertaken major efforts to identify neutral, non-prodrug inhibitors. Using a holistic analysis of all available internal data, we were able to build computational models and apply these for the selection of a lead series with the highest possibility of achieving oral bioavailability. In our design, we relied on protein structure knowledge to address potency and identified a small window of favorable physicochemical properties to balance absorption and metabolic stability. Protein structure information on the pregnane X receptor helped in overcoming a persistent cytochrome P450 3A4 induction problem. The selected compound series was optimized to a highly potent, neutral, non-prodrug thrombin inhibitor by designing, synthesizing, and testing derivatives. The resulting optimized compound, BAY1217224, has reached first clinical trials, which have confirmed the desired pharmacokinetic properties.
Organizational Affiliation:
Research and Development, Bayer AG, Pharmaceuticals, 42103 Wuppertal, Germany.