Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
Scott, J.S., Moss, T.A., Balazs, A., Barlaam, B., Breed, J., Carbajo, R.J., Chiarparin, E., Davey, P.R.J., Delpuech, O., Fawell, S., Fisher, D.I., Gagrica, S., Gangl, E.T., Grebe, T., Greenwood, R.D., Hande, S., Hatoum-Mokdad, H., Herlihy, K., Hughes, S., Hunt, T.A., Huynh, H., Janbon, S.L.M., Johnson, T., Kavanagh, S., Klinowska, T., Lawson, M., Lister, A.S., Marden, S., McGinnity, D.F., Morrow, C.J., Nissink, J.W.M., O'Donovan, D.H., Peng, B., Polanski, R., Stead, D.S., Stokes, S., Thakur, K., Throner, S.R., Tucker, M.J., Varnes, J., Wang, H., Wilson, D.M., Wu, D., Wu, Y., Yang, B., Yang, W.(2020) J Med Chem 63: 14530-14559
- PubMed: 32910656
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01163
- Primary Citation of Related Structures:
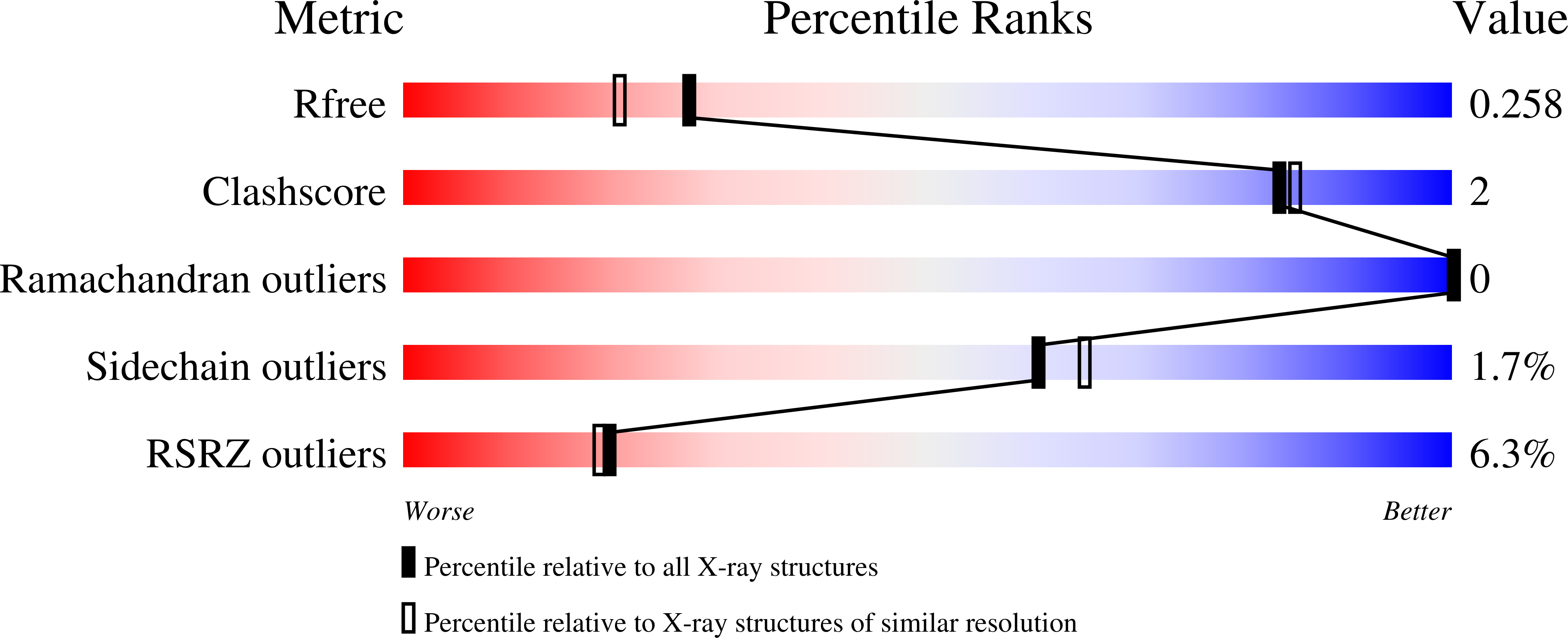
6ZOQ, 6ZOR, 6ZOS - PubMed Abstract:
Herein we report the optimization of a series of tricyclic indazoles as selective estrogen receptor degraders (SERD) and antagonists for the treatment of ER + breast cancer. Structure based design together with systematic investigation of each region of the molecular architecture led to the identification of N -[1-(3-fluoropropyl)azetidin-3-yl]-6-[(6 S ,8 R )-8-methyl-7-(2,2,2-trifluoroethyl)-6,7,8,9-tetrahydro-3 H -pyrazolo[4,3- f ]isoquinolin-6-yl]pyridin-3-amine ( 28 ). This compound was demonstrated to be a highly potent SERD that showed a pharmacological profile comparable to fulvestrant in its ability to degrade ERα in both MCF-7 and CAMA-1 cell lines. A stringent control of lipophilicity ensured that 28 had favorable physicochemical and preclinical pharmacokinetic properties for oral administration. This, combined with demonstration of potent in vivo activity in mouse xenograft models, resulted in progression of this compound, also known as AZD9833, into clinical trials.
Organizational Affiliation:
Oncology R&D, AstraZeneca, Cambridge CB4 0WG, United Kingdom.