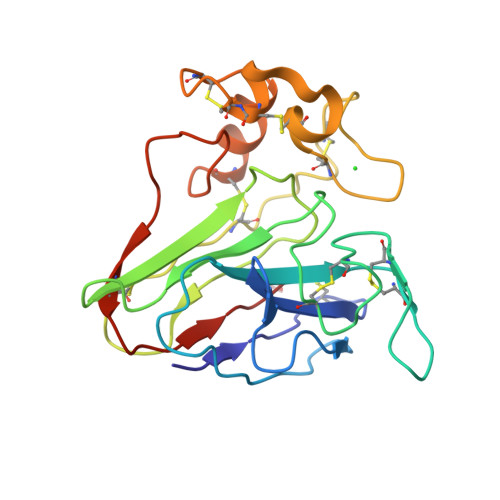
Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Blum, T.B., Housset, D., Clabbers, M.T.B., van Genderen, E., Bacia-Verloop, M., Zander, U., McCarthy, A.A., Schoehn, G., Ling, W.L., Abrahams, J.P.(2021) Acta Crystallogr D Struct Biol 77: 75-85
- PubMed: 33404527
- DOI: https://doi.org/10.1107/S2059798320014540
- Primary Citation of Related Structures:
6ZHB, 6ZHJ, 6ZHN, 6ZI8 - PubMed Abstract:
Electron diffraction allows protein structure determination when only nanosized crystals are available. Nevertheless, multiple elastic (or dynamical) scattering, which is prominent in electron diffraction, is a concern. Current methods for modeling dynamical scattering by multi-slice or Bloch wave approaches are not suitable for protein crystals because they are not designed to cope with large molecules. Here, dynamical scattering of nanocrystals of insulin, thermolysin and thaumatin was limited by collecting data from thin crystals. To accurately measure the weak diffraction signal from the few unit cells in the thin crystals, a low-noise hybrid pixel Timepix electron-counting detector was used. The remaining dynamical component was further reduced in refinement using a likelihood-based correction, which was introduced previously for analyzing electron diffraction data of small-molecule nanocrystals and was adapted here for protein crystals. The procedure is shown to notably improve the structural refinement, in one case allowing the location of solvent molecules. It also allowed refinement of the charge states of bound metal atoms, an important element in protein function, through B-factor analysis of the metal atoms and their ligands. These results clearly increase the value of macromolecular electron crystallography as a complementary structural biology technique.
Organizational Affiliation:
Department of Biology and Chemistry, Paul Scherrer Institute, 5232 Villigen, Switzerland.