New Chemical Probe Targeting Bacterial NAD Kinase.
Clement, D.A., Leseigneur, C., Gelin, M., Coelho, D., Huteau, V., Lionne, C., Labesse, G., Dussurget, O., Pochet, S.(2020) Molecules 25
- PubMed: 33105870
- DOI: https://doi.org/10.3390/molecules25214893
- Primary Citation of Related Structures:
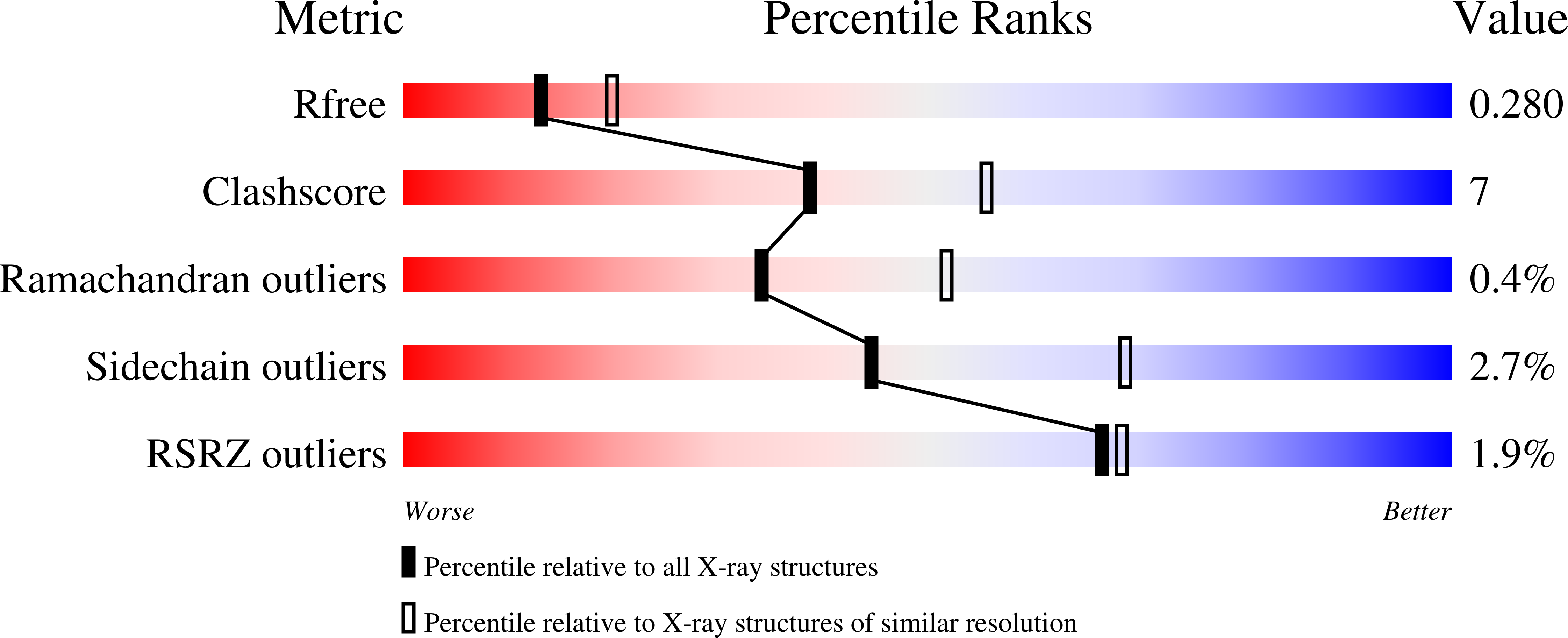
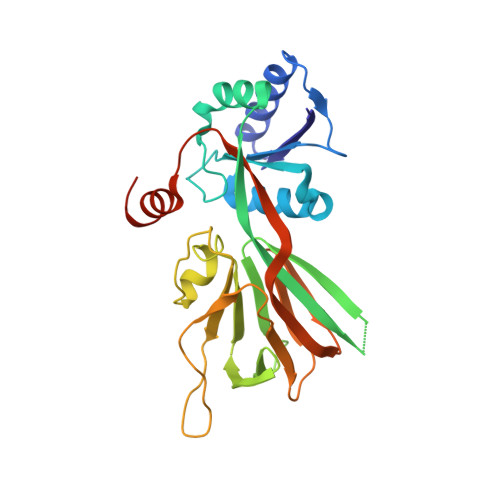
6Z61, 6Z64, 6Z65 - PubMed Abstract:
Nicotinamide adenine dinucleotide (NAD) kinases are essential and ubiquitous enzymes involved in the tight regulation of NAD/nicotinamide adenine dinucleotide phosphate (NADP) levels in many metabolic pathways. Consequently, they represent promising therapeutic targets in cancer and antibacterial treatments. We previously reported diadenosine derivatives as NAD kinase inhibitors with bactericidal activities on Staphylococcus aureus . Among them, one compound (namely NKI1 ) was found effective in vivo in a mouse infection model. With the aim to gain detailed knowledge about the selectivity and mechanism of action of this lead compound, we planned to develop a chemical probe that could be used in affinity-based chemoproteomic approaches. Here, we describe the first functionalized chemical probe targeting a bacterial NAD kinase. Aminoalkyl functional groups were introduced on NKI1 for further covalent coupling to an activated Sepharose TM matrix. Inhibitory properties of functionalized NKI1 derivatives together with X-ray characterization of their complexes with the NAD kinase led to identify candidate compounds that are amenable to covalent coupling to a matrix.
Organizational Affiliation:
Institut Pasteur, Unité de Chimie et Biocatalyse, UMR3523 CNRS, 75015 Paris, France.