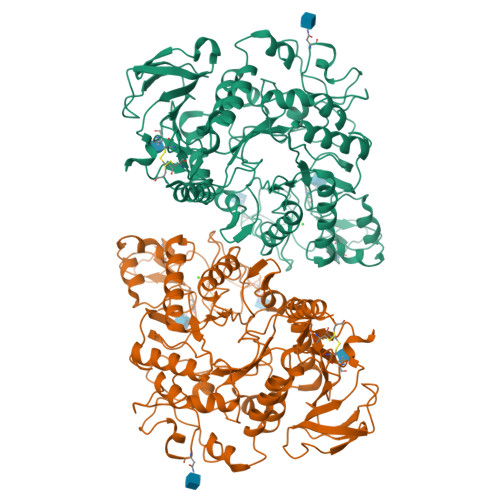
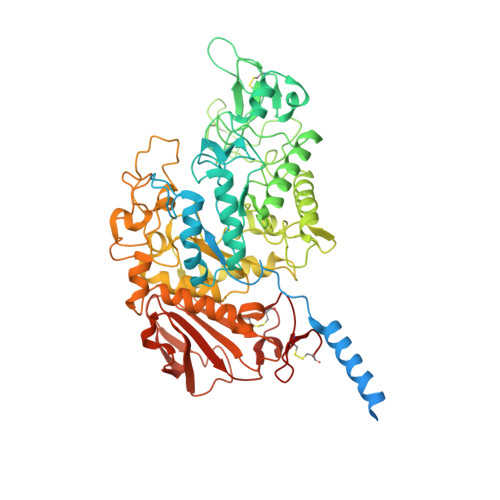
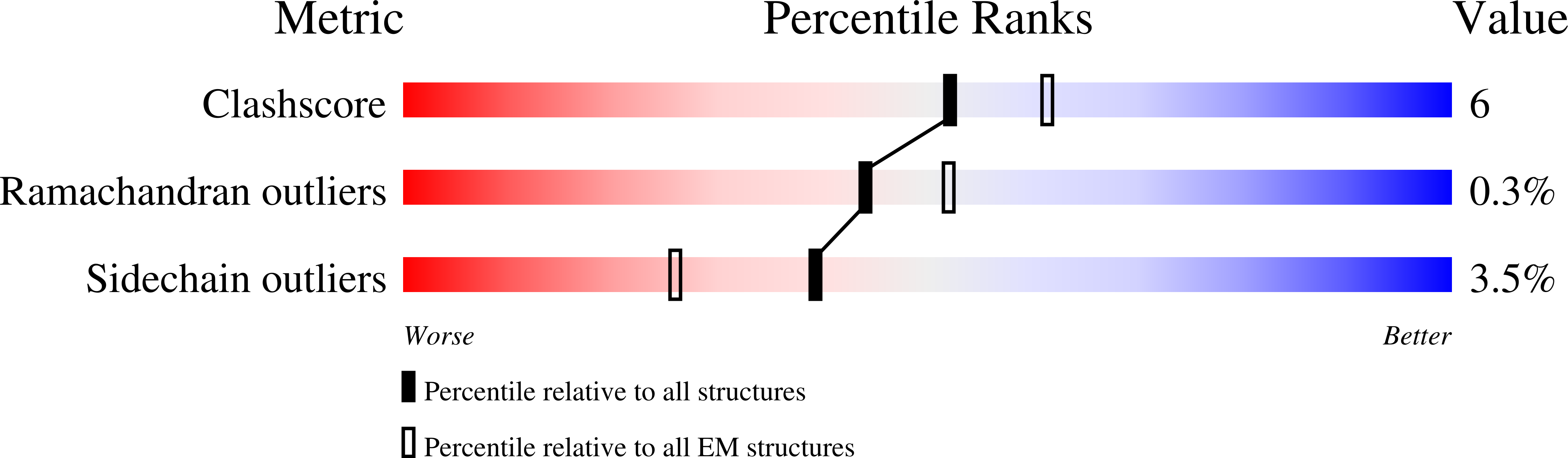Structural basis for amino acid exchange by a human heteromeric amino acid transporter.
Wu, D., Grund, T.N., Welsch, S., Mills, D.J., Michel, M., Safarian, S., Michel, H.(2020) Proc Natl Acad Sci U S A 117: 21281-21287
- PubMed: 32817565
- DOI: https://doi.org/10.1073/pnas.2008111117
- Primary Citation of Related Structures:
6YUP, 6YUZ, 6YV1 - PubMed Abstract:
Heteromeric amino acid transporters (HATs) comprise a group of membrane proteins that belong to the solute carrier (SLC) superfamily. They are formed by two different protein components: a light chain subunit from an SLC7 family member and a heavy chain subunit from the SLC3 family. The light chain constitutes the transport subunit whereas the heavy chain mediates trafficking to the plasma membrane and maturation of the functional complex. Mutation, malfunction, and dysregulation of HATs are associated with a wide range of pathologies or represent the direct cause of inherited and acquired disorders. Here we report the cryogenic electron microscopy structure of the neutral and basic amino acid transport complex (b [0,+] AT1-rBAT) which reveals a heterotetrameric protein assembly composed of two heavy and light chain subunits, respectively. The previously uncharacterized interaction between two HAT units is mediated via dimerization of the heavy chain subunits and does not include participation of the light chain subunits. The b (0,+) AT1 transporter adopts a LeuT fold and is captured in an inward-facing conformation. We identify an amino-acid-binding pocket that is formed by transmembrane helices 1, 6, and 10 and conserved among SLC7 transporters.
Organizational Affiliation:
Department of Molecular Membrane Biology, Max Planck Institute of Biophysics, D-60438 Frankfurt am Main, Germany.