The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
Romero-Romero, S., Costas, M., Silva Manzano, D.A., Kordes, S., Rojas-Ortega, E., Tapia, C., Guerra, Y., Shanmugaratnam, S., Rodriguez-Romero, A., Baker, D., Hocker, B., Fernandez-Velasco, D.A.(2021) J Mol Biol 433: 167153-167153
- PubMed: 34271011
- DOI: https://doi.org/10.1016/j.jmb.2021.167153
- Primary Citation of Related Structures:
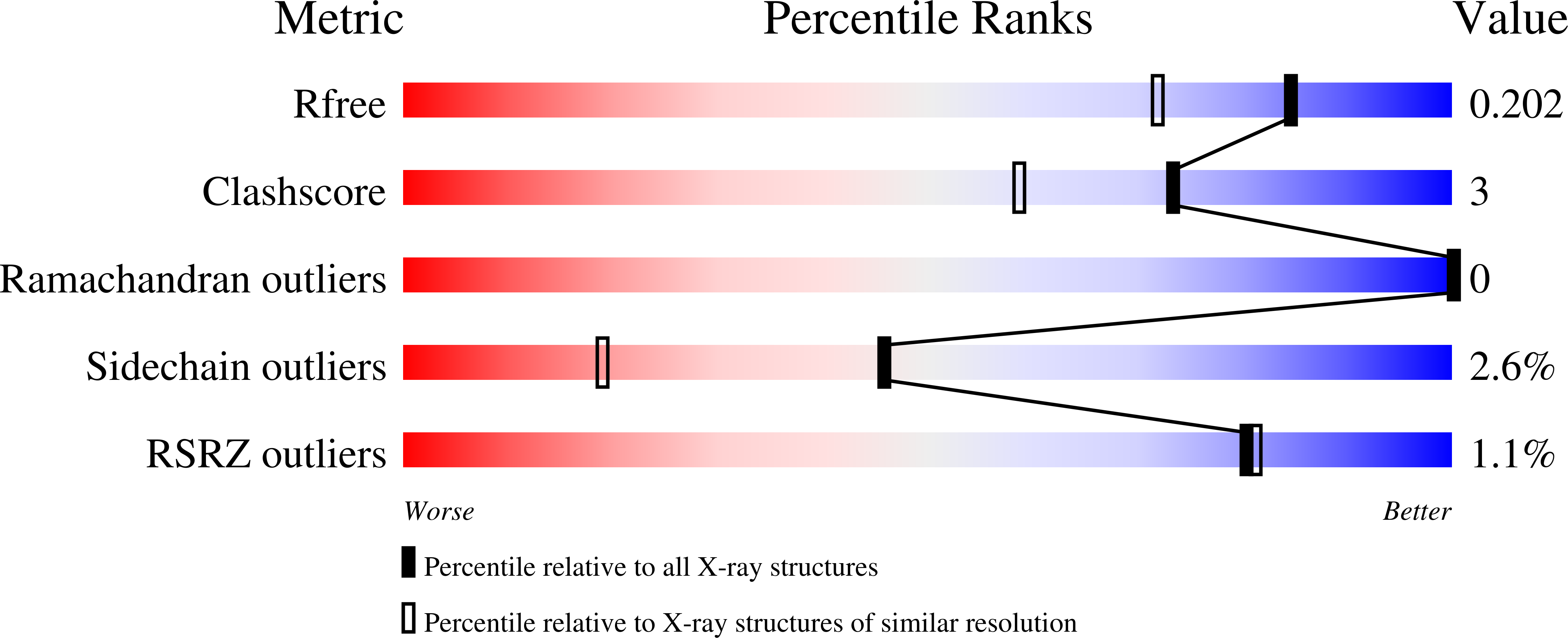
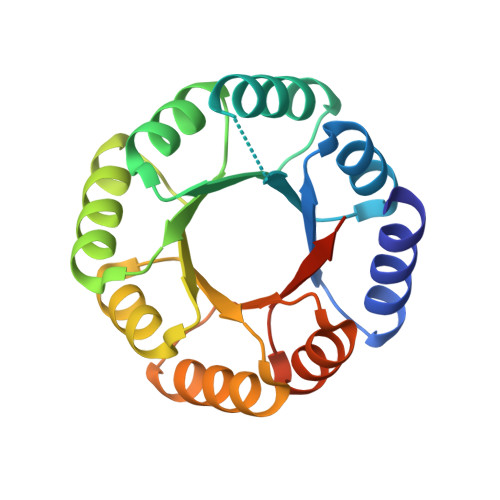
6YQX, 6YQY, 6Z2I - PubMed Abstract:
The ability to design stable proteins with custom-made functions is a major goal in biochemistry with practical relevance for our environment and society. Understanding and manipulating protein stability provide crucial information on the molecular determinants that modulate structure and stability, and expand the applications of de novo proteins. Since the (β/⍺) 8 -barrel or TIM-barrel fold is one of the most common functional scaffolds, in this work we designed a collection of stable de novo TIM barrels (DeNovoTIMs), using a computational fixed-backbone and modular approach based on improved hydrophobic packing of sTIM11, the first validated de novo TIM barrel, and subjected them to a thorough folding analysis. DeNovoTIMs navigate a region of the stability landscape previously uncharted by natural TIM barrels, with variations spanning 60 degrees in melting temperature and 22 kcal per mol in conformational stability throughout the designs. Significant non-additive or epistatic effects were observed when stabilizing mutations from different regions of the barrel were combined. The molecular basis of epistasis in DeNovoTIMs appears to be related to the extension of the hydrophobic cores. This study is an important step towards the fine-tuned modulation of protein stability by design.
Organizational Affiliation:
Laboratorio de Fisicoquímica e Ingeniería de Proteínas, Departamento de Bioquímica, Facultad de Medicina, Universidad Nacional Autónoma de México, 04510 Mexico City, Mexico; Department of Biochemistry, University of Bayreuth, 95447 Bayreuth, Germany.