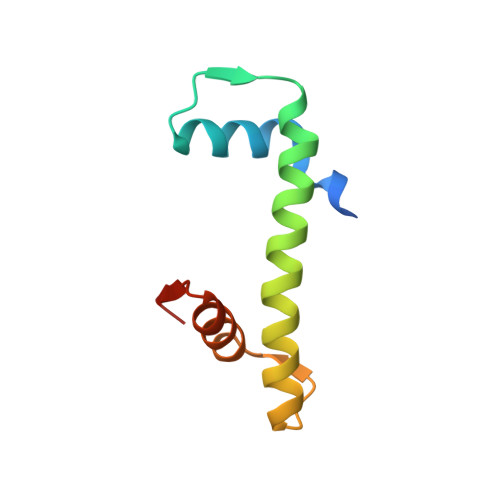
Chaperoning of the histone octamer by the acidic domain of DNA repair factor APLF.
Corbeski, I., Guo, X., Eckhardt, B.V., Fasci, D., Wiegant, W., Graewert, M.A., Vreeken, K., Wienk, H., Svergun, D.I., Heck, A.J.R., van Attikum, H., Boelens, R., Sixma, T.K., Mattiroli, F., van Ingen, H.(2022) Sci Adv 8: eabo0517-eabo0517
- PubMed: 35895815
- DOI: https://doi.org/10.1126/sciadv.abo0517
- Primary Citation of Related Structures:
6YN1 - PubMed Abstract:
Nucleosome assembly requires the coordinated deposition of histone complexes H3-H4 and H2A-H2B to form a histone octamer on DNA. In the current paradigm, specific histone chaperones guide the deposition of first H3-H4 and then H2A-H2B. Here, we show that the acidic domain of DNA repair factor APLF (APLF AD ) can assemble the histone octamer in a single step and deposit it on DNA to form nucleosomes. The crystal structure of the APLF AD -histone octamer complex shows that APLF AD tethers the histones in their nucleosomal conformation. Mutations of key aromatic anchor residues in APLF AD affect chaperone activity in vitro and in cells. Together, we propose that chaperoning of the histone octamer is a mechanism for histone chaperone function at sites where chromatin is temporarily disrupted.
- NMR Spectroscopy, Bijvoet Centre for Biomolecular Research, Utrecht University, Padualaan 8, 3584 CH Utrecht, Netherlands.
Organizational Affiliation: