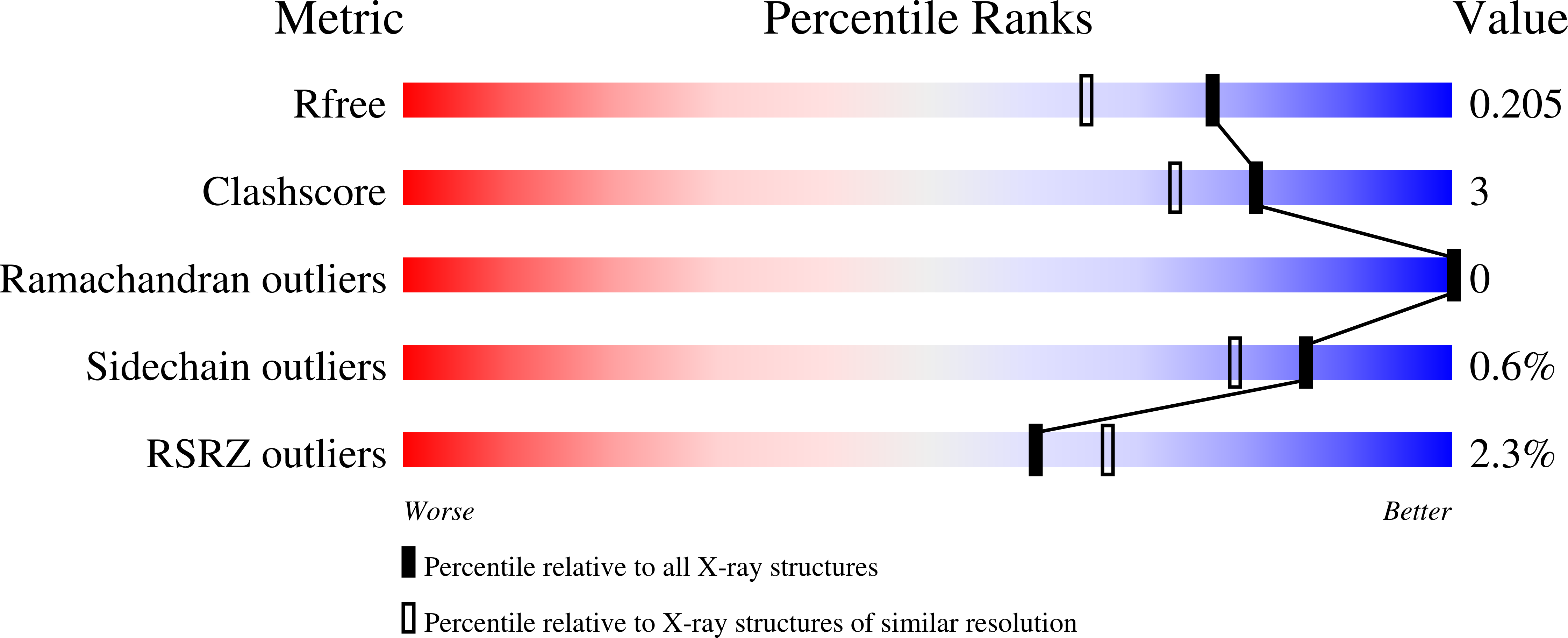
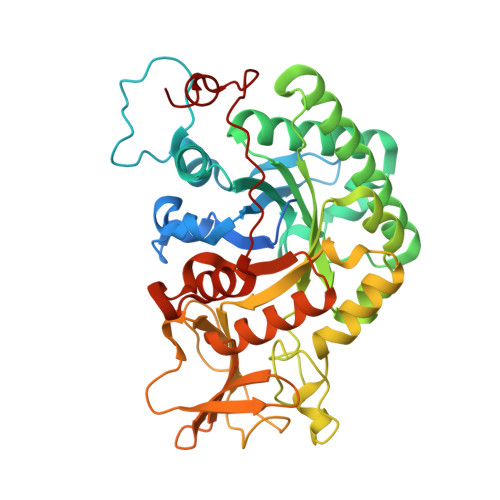
Structural inspection and protein motions modelling of a fungal glycoside hydrolase family 18 chitinase by crystallography depicts a dynamic enzymatic mechanism
Jimenez-Ortega, E., Kidibule, P.E., Fernandez-Lobato, M., Sanz-Aparicio, J.(2021) Comput Struct Biotechnol J 19: 5466-5478