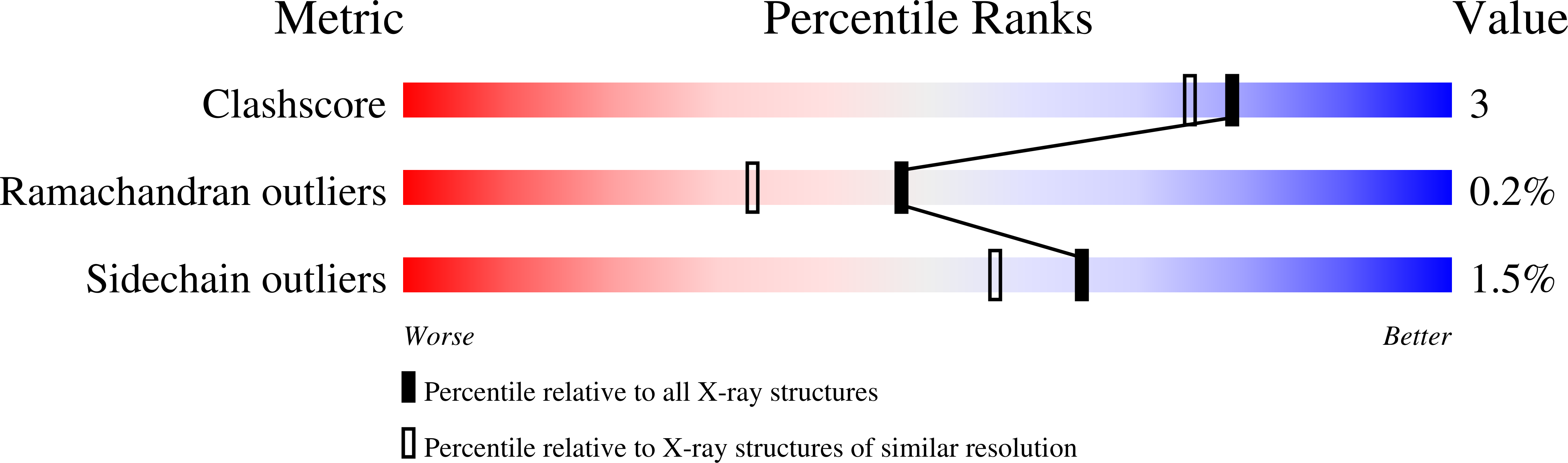Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Ellis, E.S., Hinchen, D.J., Bleem, A., Bu, L., Mallinson, S.J.B., Allen, M.D., Streit, B.R., Machovina, M.M., Doolin, Q.V., Michener, W.E., Johnson, C.W., Knott, B.C., Beckham, G.T., McGeehan, J.E., DuBois, J.L.(2021) JACS Au 1: 252-261
- PubMed: 34467290
- DOI: https://doi.org/10.1021/jacsau.0c00103
- Primary Citation of Related Structures:
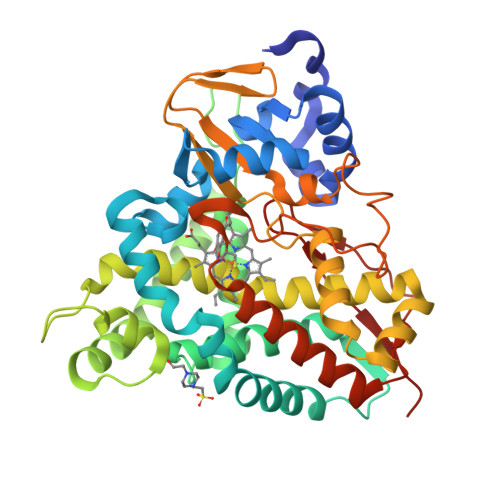
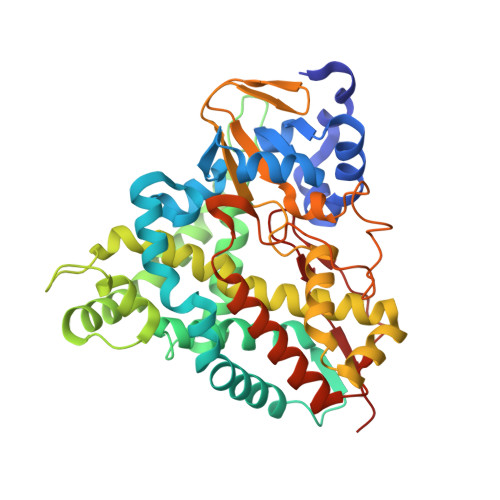
6YCH, 6YCI, 6YCJ, 6YCK, 6YCL, 6YCM, 6YCN, 6YCO, 6YCP, 6YCT - PubMed Abstract:
Biological funneling of lignin-derived aromatic compounds is a promising approach for valorizing its catalytic depolymerization products. Industrial processes for aromatic bioconversion will require efficient enzymes for key reactions, including demethylation of O -methoxy-aryl groups, an essential and often rate-limiting step. The recently characterized GcoAB cytochrome P450 system comprises a coupled monoxygenase (GcoA) and reductase (GcoB) that catalyzes oxidative demethylation of the O- methoxy-aryl group in guaiacol. Here, we evaluate a series of engineered GcoA variants for their ability to demethylate o -and p -vanillin, which are abundant lignin depolymerization products. Two rationally designed, single amino acid substitutions, F169S and T296S, are required to convert GcoA into an efficient catalyst toward the o - and p -isomers of vanillin, respectively. Gain-of-function in each case is explained in light of an extensive series of enzyme-ligand structures, kinetic data, and molecular dynamics simulations. Using strains of Pseudomonas putida KT2440 already optimized for p -vanillin production from ferulate, we demonstrate demethylation by the T296S variant in vivo . This work expands the known aromatic O- demethylation capacity of cytochrome P450 enzymes toward important lignin-derived aromatic monomers.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Montana State University, 103 Chemistry and Biochemistry Building, PO Box 173400, Bozeman, Montana 59717, United States.