Reconstitution and Structural Analysis of a HECT Ligase-Ubiquitin Complex via an Activity-Based Probe.
Nair, R.M., Seenivasan, A., Liu, B., Chen, D., Lowe, E.D., Lorenz, S.(2021) ACS Chem Biol 16: 1615-1621
- PubMed: 34403242
- DOI: https://doi.org/10.1021/acschembio.1c00433
- Primary Citation of Related Structures:
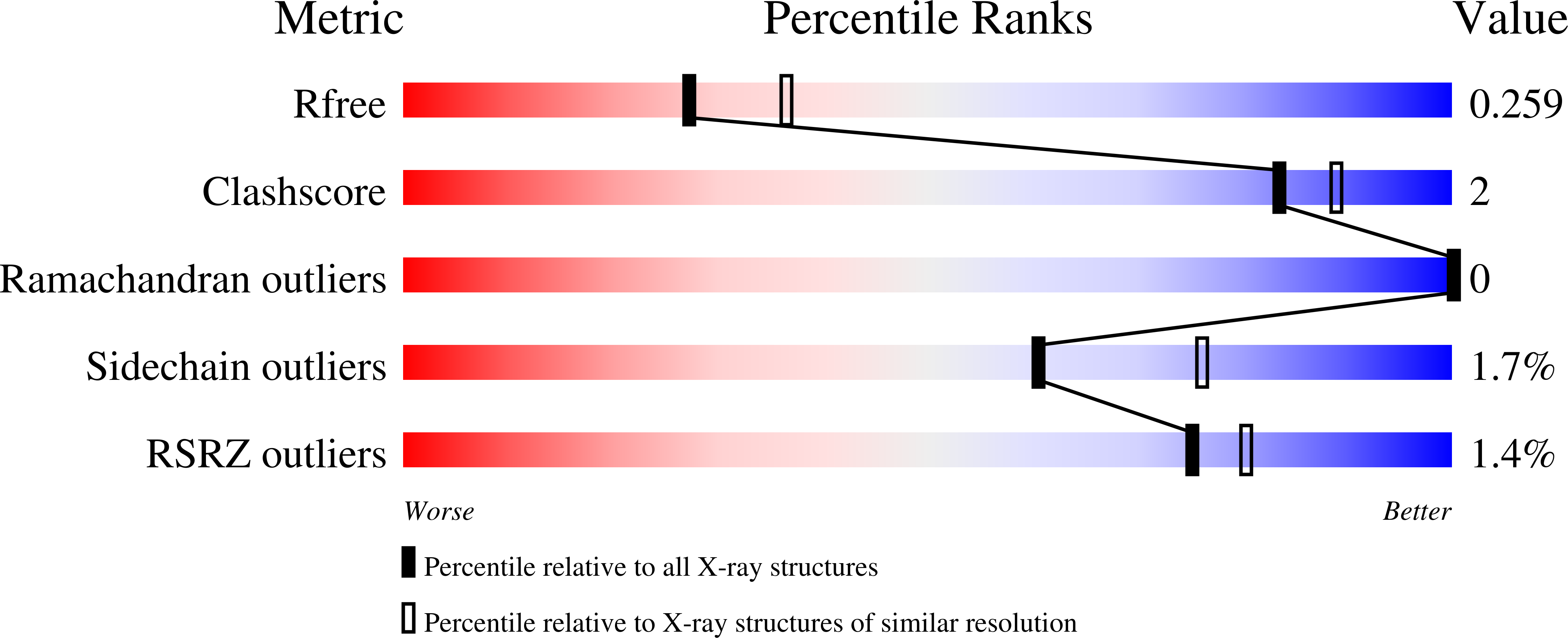
6XZ1 - PubMed Abstract:
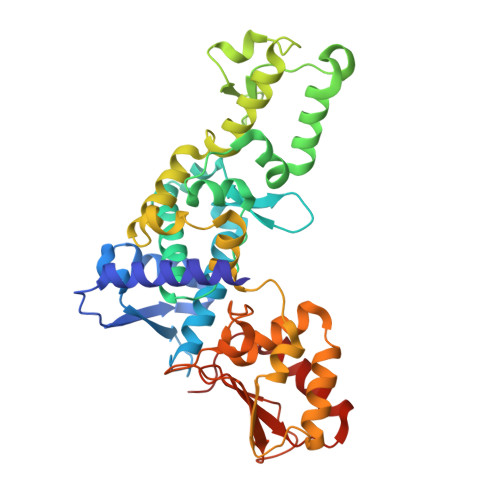
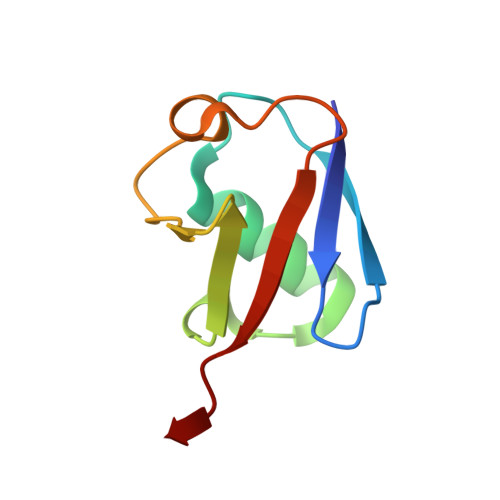
Ubiquitin activity-based probes have proven invaluable in elucidating structural mechanisms in the ubiquitin system by stabilizing transient macromolecular complexes of deubiquitinases, ubiquitin-activating enzymes, and the assemblies of ubiquitin-conjugating enzymes with ubiquitin ligases of the RING-Between-RING and RING-Cysteine-Relay families. Here, we demonstrate that an activity-based probe, ubiquitin-propargylamine, allows for the preparative reconstitution and structural analysis of the interactions between ubiquitin and certain HECT ligases. We present a crystal structure of the ubiquitin-linked HECT domain of HUWE1 that defines a catalytically critical conformation of the C-terminal tail of the ligase for the transfer of ubiquitin to an acceptor protein. Moreover, we observe that ubiquitin-propargylamine displays selectivity among HECT domains, thus corroborating the notion that activity-based probes may provide entry points for the development of specific, active site-directed inhibitors and reporters of HECT ligase activities.
Organizational Affiliation:
Rudolf Virchow Center for Integrative and Translational Bioimaging, University of Würzburg, 97080 Würzburg, Germany.