Nanobodies Targeting Prostate-Specific Membrane Antigen for the Imaging and Therapy of Prostate Cancer.
Rosenfeld, L., Sananes, A., Zur, Y., Cohen, S., Dhara, K., Gelkop, S., Ben Zeev, E., Shahar, A., Lobel, L., Akabayov, B., Arbely, E., Papo, N.(2020) J Med Chem 63: 7601-7615
- PubMed: 32442375
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00418
- Primary Citation of Related Structures:
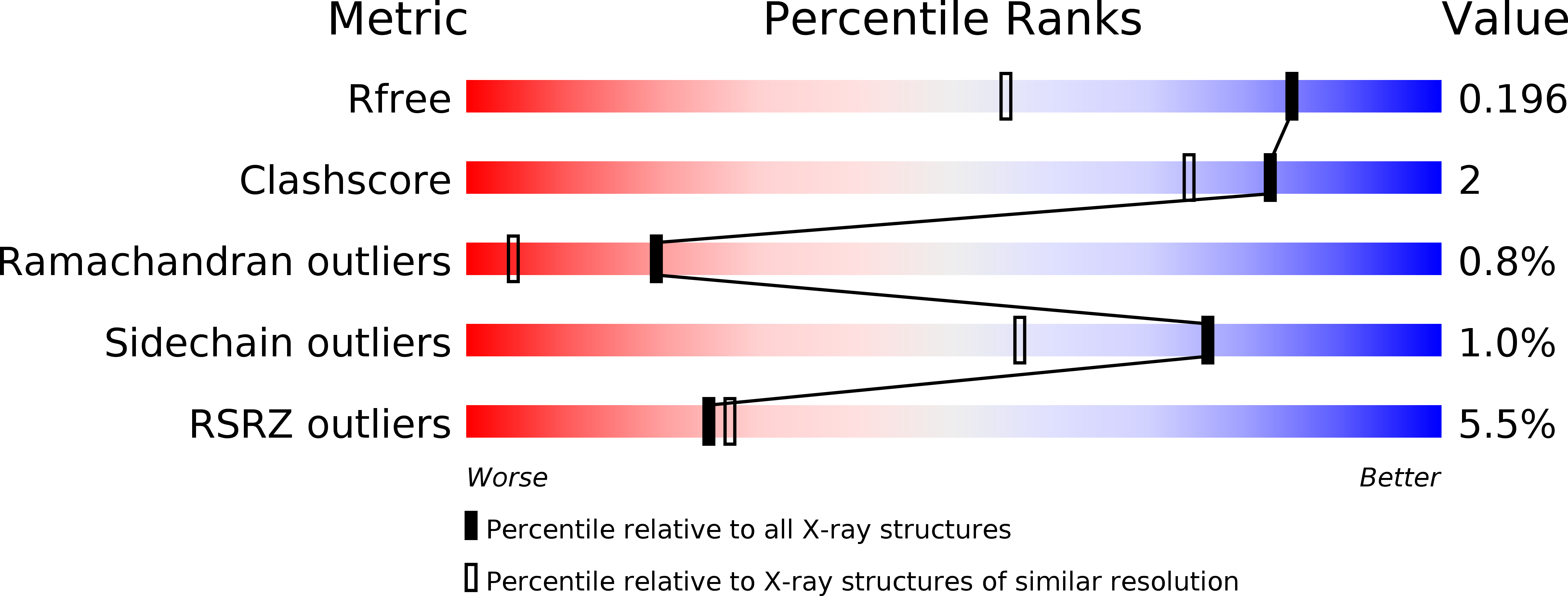
6XXN, 6XXO, 6XXP - PubMed Abstract:
The repertoire of methods for the detection and chemotherapeutic treatment of prostate cancer (PCa) is currently limited. Prostate-specific membrane antigen (PSMA) is overexpressed in PCa tumors and can be exploited for both imaging and drug delivery. We developed and characterized four nanobodies that present tight and specific binding and internalization into PSMA + cells and that accumulate specifically in PSMA + tumors. We then conjugated one of these nanobodies to the cytotoxic drug doxorubicin, and we show that the conjugate internalizes specifically into PSMA + cells, where the drug is released and induces cytotoxic activity. In vivo studies show that the extent of tumor growth inhibition is similar when mice are treated with commercial doxorubicin and with a 42-fold lower amount of the nanobody-conjugated doxorubicin, attesting to the efficacy of the conjugated drug. These data highlight nanobodies as promising agents for the imaging of PCa tumors and for the targeted delivery of chemotherapeutic drugs.
Organizational Affiliation:
Avram and Stella Goldstein-Goren Department of Biotechnology Engineering and the National Institute of Biotechnology in the Negev, Ben-Gurion University of the Negev, Beer-Sheva 8410501, Israel.