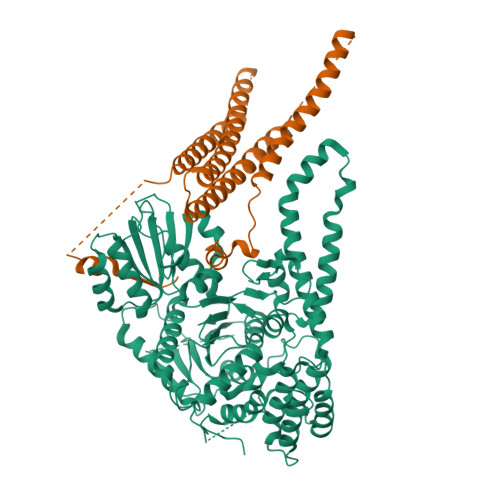
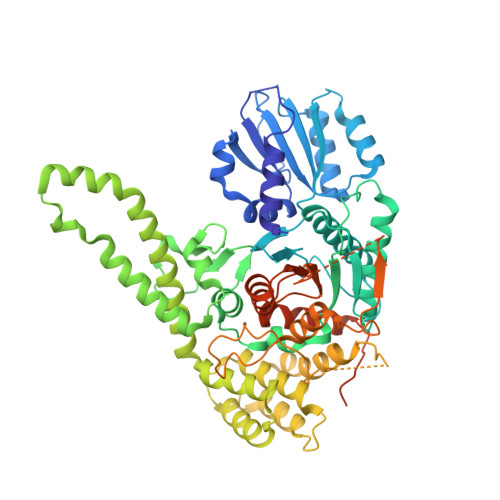
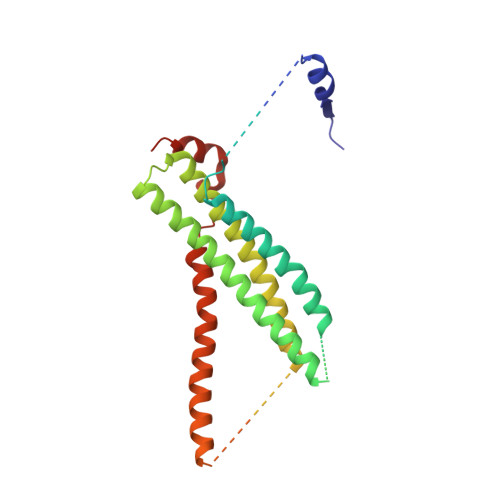
The Sec1/Munc18 protein Vps45 holds the Qa-SNARE Tlg2 in an open conformation.
Eisemann, T.J., Allen, F., Lau, K., Shimamura, G.R., Jeffrey, P.D., Hughson, F.M.(2020) Elife 9
- PubMed: 32804076
- DOI: https://doi.org/10.7554/eLife.60724
- Primary Citation of Related Structures:
6XJL, 6XM1, 6XMD - PubMed Abstract:
Fusion of intracellular trafficking vesicles is mediated by the assembly of SNARE proteins into membrane-bridging complexes. SNARE-mediated membrane fusion requires Sec1/Munc18-family (SM) proteins, SNARE chaperones that can function as templates to catalyze SNARE complex assembly. Paradoxically, the SM protein Munc18-1 traps the Qa-SNARE protein syntaxin-1 in an autoinhibited closed conformation. Here we present the structure of a second SM-Qa-SNARE complex, Vps45-Tlg2. Strikingly, Vps45 holds Tlg2 in an open conformation, with its SNARE motif disengaged from its Habc domain and its linker region unfolded. The domain 3a helical hairpin of Vps45 is unfurled, exposing the presumptive R-SNARE binding site to allow template complex formation. Although Tlg2 has a pronounced tendency to form homo-tetramers, Vps45 can rescue Tlg2 tetramers into stoichiometric Vps45-Tlg2 complexes. Our findings demonstrate that SM proteins can engage Qa-SNAREs using at least two different modes, one in which the SNARE is closed and one in which it is open.
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Princeton, United States.