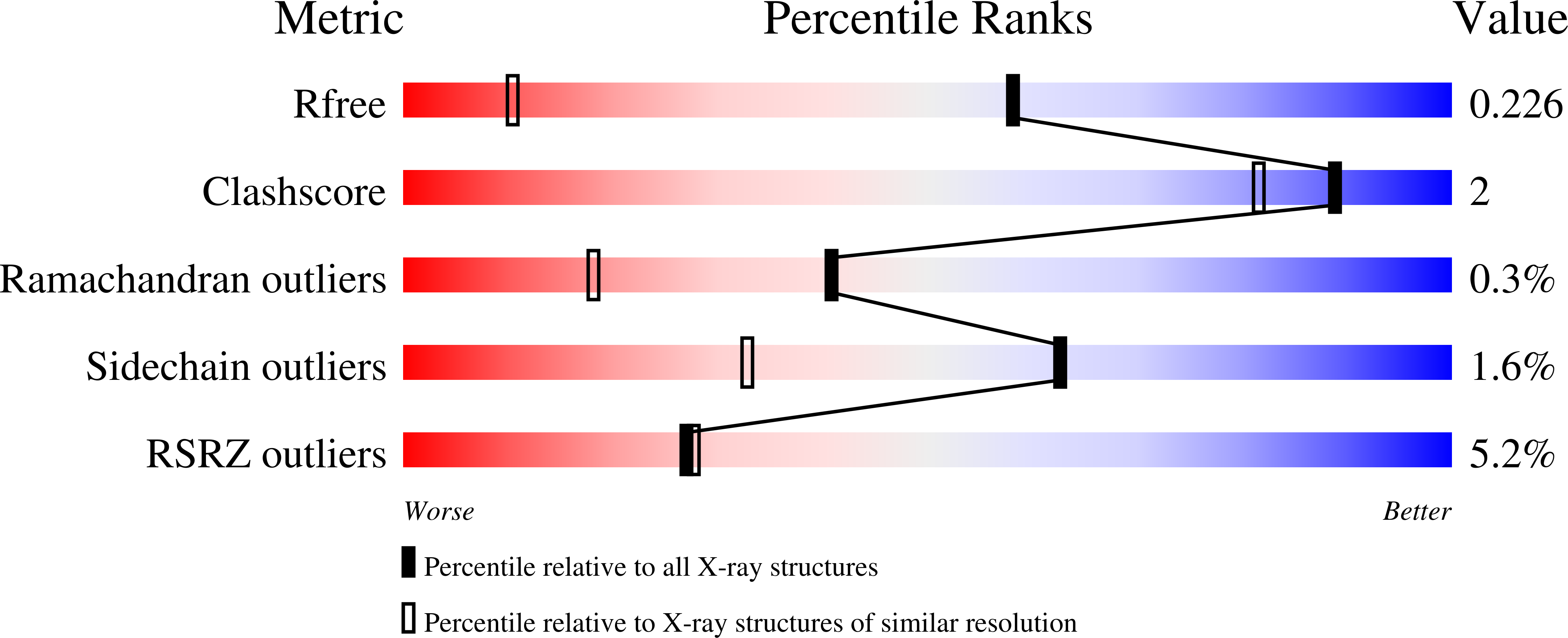
Series of Novel and Highly Potent Cyclic Peptide PCSK9 Inhibitors Derived from an mRNA Display Screen and Optimized via Structure-Based Design.
Alleyne, C., Amin, R.P., Bhatt, B., Bianchi, E., Blain, J.C., Boyer, N., Branca, D., Embrey, M.W., Ha, S.N., Jette, K., Johns, D.G., Kerekes, A.D., Koeplinger, K.A., LaPlaca, D., Li, N., Murphy, B., Orth, P., Ricardo, A., Salowe, S., Seyb, K., Shahripour, A., Stringer, J.R., Sun, Y., Tracy, R., Wu, C., Xiong, Y., Youm, H., Zokian, H.J., Tucker, T.J.(2020) J Med Chem 63: 13796-13824
- PubMed: 33170686
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01084
- Primary Citation of Related Structures:
6XIB, 6XIC, 6XID, 6XIE, 6XIF - PubMed Abstract:
Proprotein convertase subtilisin-like/kexin type 9 (PCSK9) is a key regulator of plasma LDL-cholesterol (LDL-C) and a clinically validated target for the treatment of hypercholesterolemia and coronary artery disease. In this paper, we describe a series of novel cyclic peptides derived from an mRNA display screen which inhibit the protein-protein interaction between PCSK9 and LDLR. Using a structure-based drug design approach, we were able to modify our original screening lead 2 to optimize the potency and metabolic stability and minimize the molecular weight to provide novel bicyclic next-generation PCSK9 inhibitor peptides such as 78 . These next-generation peptides serve as a critical foundation for continued exploration of potential oral, once-a-day PCSK9 therapeutics for the treatment of cardiovascular disease.
Organizational Affiliation:
Discovery Pharmaceutical Sciences, Merck & Company, Inc., 2000 Galloping Hill Road, Kenilworth, New Jersey 07033, United States.