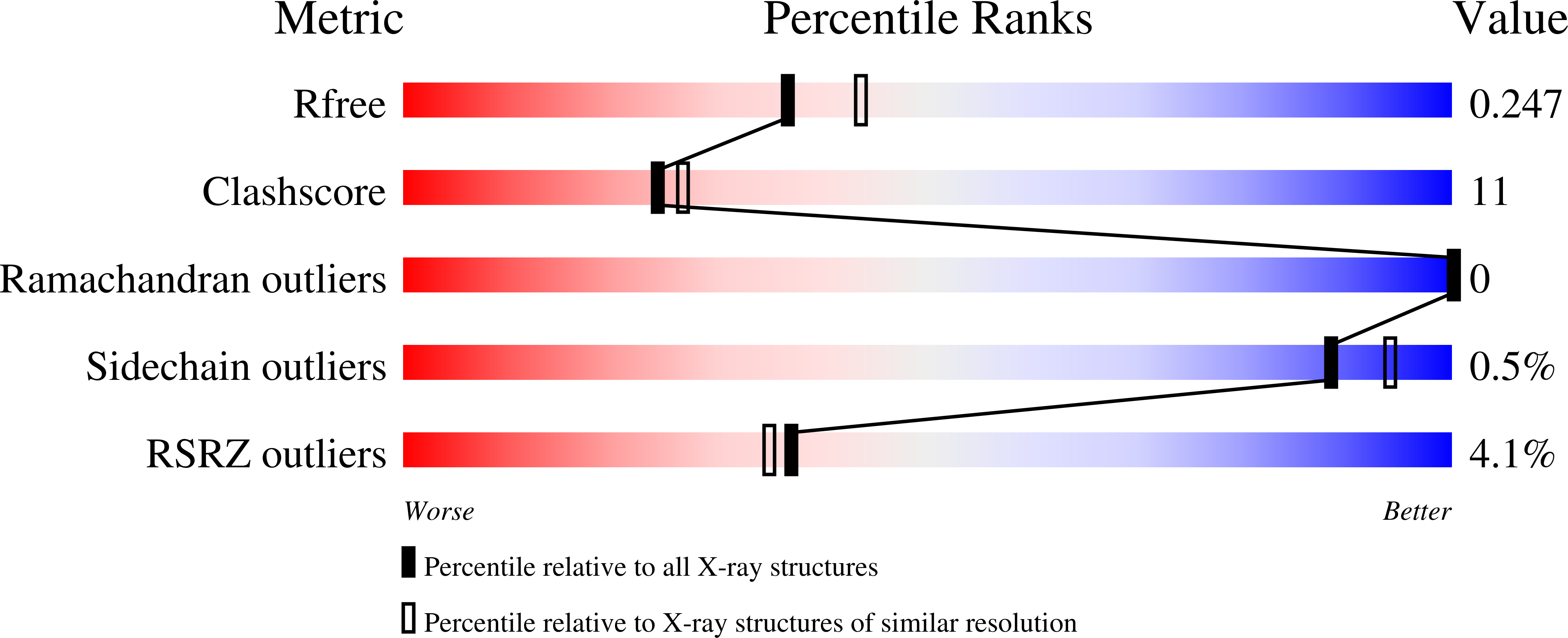
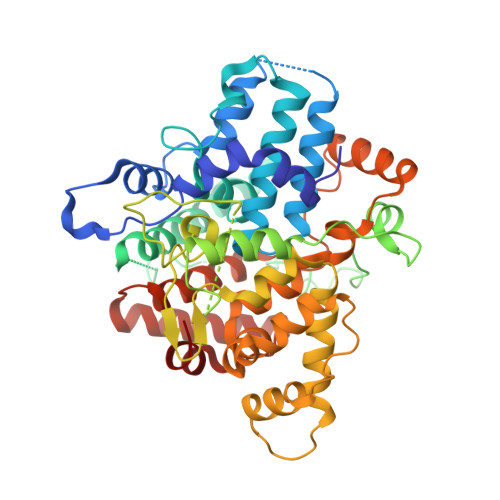
A pentameric protein ring with novel architecture is required for herpesviral packaging.
Didychuk, A.L., Gates, S.N., Gardner, M.R., Strong, L.M., Martin, A., Glaunsinger, B.A.(2021) Elife 10
- PubMed: 33554858
- DOI: https://doi.org/10.7554/eLife.62261
- Primary Citation of Related Structures:
6XF9, 6XFA - PubMed Abstract:
Genome packaging in large double-stranded DNA viruses requires a powerful molecular motor to force the viral genome into nascent capsids, which involves essential accessory factors that are poorly understood. Here, we present structures of two such accessory factors from the oncogenic herpesviruses Kaposi's sarcoma-associated herpesvirus (KSHV; ORF68) and Epstein-Barr virus (EBV; BFLF1). These homologous proteins form highly similar homopentameric rings with a positively charged central channel that binds double-stranded DNA. Mutation of individual positively charged residues within but not outside the channel ablates DNA binding, and in the context of KSHV infection, these mutants fail to package the viral genome or produce progeny virions. Thus, we propose a model in which ORF68 facilitates the transfer of newly replicated viral genomes to the packaging motor.
Organizational Affiliation:
Department of Plant and Microbial Biology, University of California, Berkeley, Berkeley, United States.