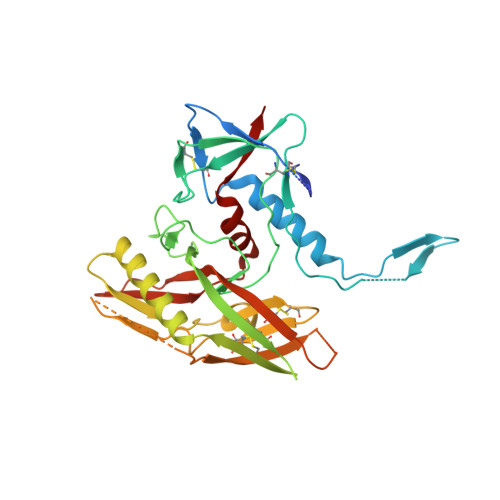
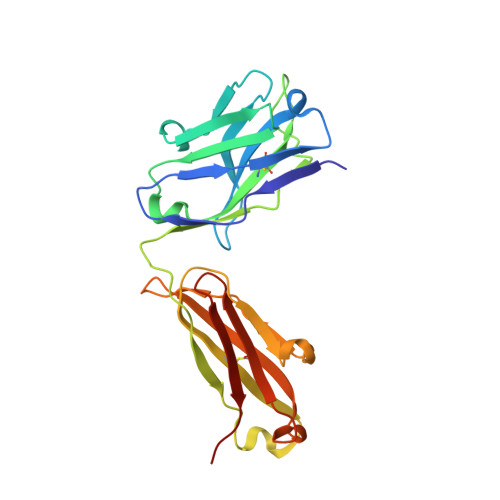
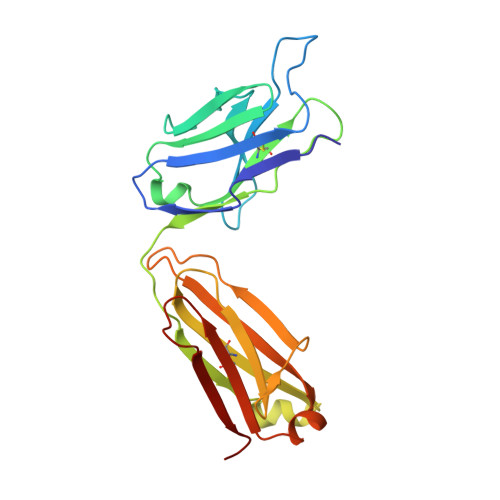
Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth.
Roark, R.S., Li, H., Williams, W.B., Chug, H., Mason, R.D., Gorman, J., Wang, S., Lee, F.H., Rando, J., Bonsignori, M., Hwang, K.K., Saunders, K.O., Wiehe, K., Moody, M.A., Hraber, P.T., Wagh, K., Giorgi, E.E., Russell, R.M., Bibollet-Ruche, F., Liu, W., Connell, J., Smith, A.G., DeVoto, J., Murphy, A.I., Smith, J., Ding, W., Zhao, C., Chohan, N., Okumura, M., Rosario, C., Ding, Y., Lindemuth, E., Bauer, A.M., Bar, K.J., Ambrozak, D., Chao, C.W., Chuang, G.Y., Geng, H., Lin, B.C., Louder, M.K., Nguyen, R., Zhang, B., Lewis, M.G., Raymond, D.D., Doria-Rose, N.A., Schramm, C.A., Douek, D.C., Roederer, M., Kepler, T.B., Kelsoe, G., Mascola, J.R., Kwong, P.D., Korber, B.T., Harrison, S.C., Haynes, B.F., Hahn, B.H., Shaw, G.M.(2021) Science 371
- PubMed: 33214287
- DOI: https://doi.org/10.1126/science.abd2638
- Primary Citation of Related Structures:
6XCJ, 6XRT - PubMed Abstract:
Neutralizing antibodies elicited by HIV-1 coevolve with viral envelope proteins (Env) in distinctive patterns, in some cases acquiring substantial breadth. We report that primary HIV-1 envelope proteins-when expressed by simian-human immunodeficiency viruses in rhesus macaques-elicited patterns of Env-antibody coevolution very similar to those in humans, including conserved immunogenetic, structural, and chemical solutions to epitope recognition and precise Env-amino acid substitutions, insertions, and deletions leading to virus persistence. The structure of one rhesus antibody, capable of neutralizing 49% of a 208-strain panel, revealed a V2 apex mode of recognition like that of human broadly neutralizing antibodies (bNAbs) PGT145 and PCT64-35S. Another rhesus antibody bound the CD4 binding site by CD4 mimicry, mirroring human bNAbs 8ANC131, CH235, and VRC01. Virus-antibody coevolution in macaques can thus recapitulate developmental features of human bNAbs, thereby guiding HIV-1 immunogen design.
Organizational Affiliation:
Departments of Medicine and Microbiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.