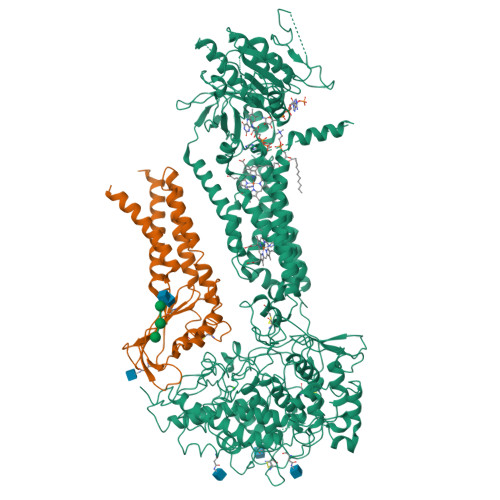
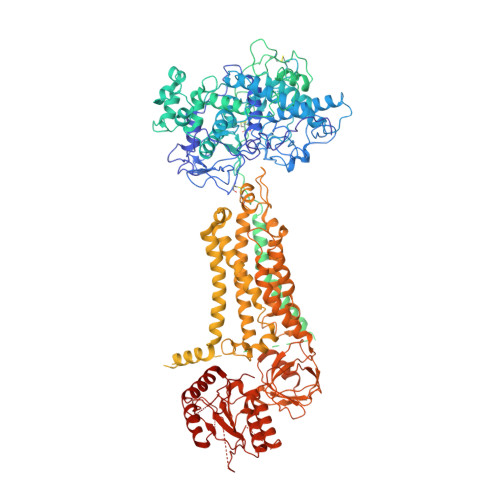
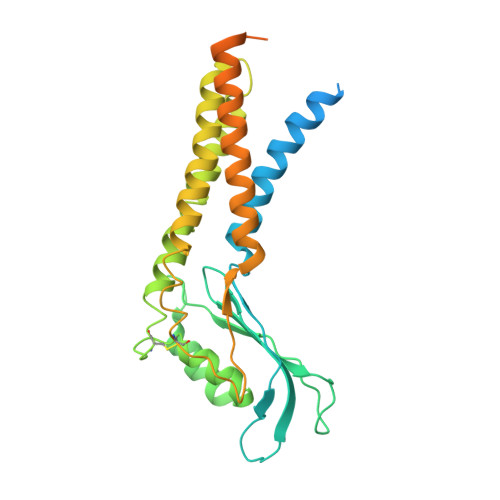
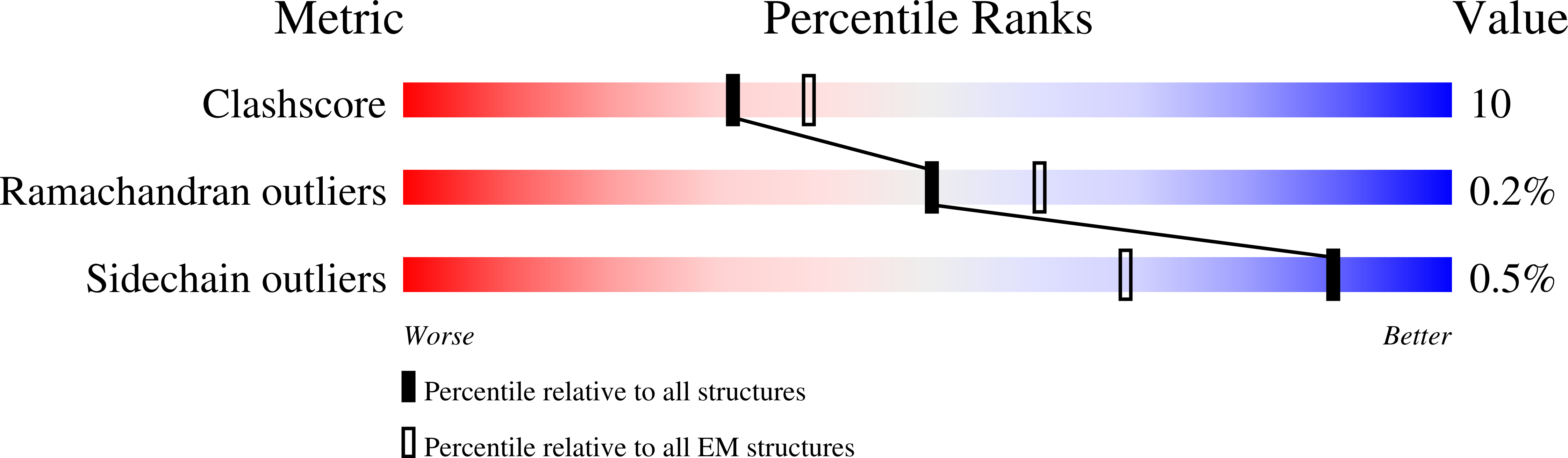Structures of mouse DUOX1-DUOXA1 provide mechanistic insights into enzyme activation and regulation.
Sun, J.(2020) Nat Struct Mol Biol 27: 1086-1093
- PubMed: 32929281
- DOI: https://doi.org/10.1038/s41594-020-0501-x
- Primary Citation of Related Structures:
6WXR, 6WXU, 6WXV - PubMed Abstract:
DUOX1, an NADPH oxidase family member, catalyzes the production of hydrogen peroxide. DUOX1 is expressed in various tissues, including the thyroid and respiratory tract, and plays a crucial role in processes such as thyroid hormone biosynthesis and innate host defense. DUOX1 co-assembles with its maturation factor DUOXA1 to form an active enzyme complex. However, the molecular mechanisms for activation and regulation of DUOX1 remain mostly unclear. Here, I present cryo-EM structures of the mammalian DUOX1-DUOXA1 complex, in the absence and presence of substrate NADPH, as well as DUOX1-DUOXA1 in an unexpected dimer-of-dimers configuration. These structures reveal atomic details of the DUOX1-DUOXA1 interaction, a lipid-mediated NADPH-binding pocket and the electron transfer path. Furthermore, biochemical and structural analyses indicate that the dimer-of-dimers configuration represents an inactive state of DUOX1-DUOXA1, suggesting an oligomerization-dependent regulatory mechanism. Together, my work provides structural bases for DUOX1-DUOXA1 activation and regulation.
Organizational Affiliation:
Department of Structural Biology, St Jude Children's Research Hospital, Memphis, TN, USA. ji.sun@stjude.org.