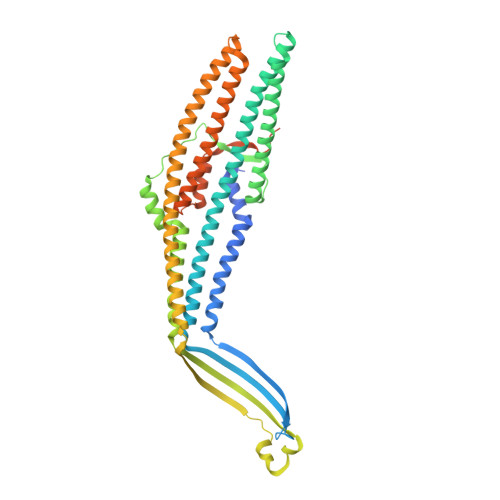
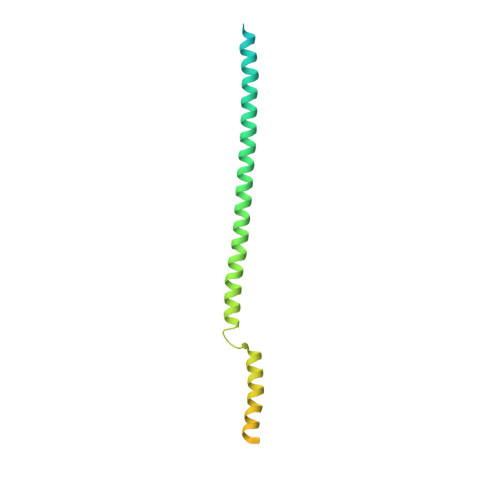
Colicin E1 opens its hinge to plug TolC.
Budiardjo, S.J., Stevens, J.J., Calkins, A.L., Ikujuni, A.P., Wimalasena, V.K., Firlar, E., Case, D.A., Biteen, J.S., Kaelber, J.T., Slusky, J.S.G.(2022) Elife 11
- PubMed: 35199644
- DOI: https://doi.org/10.7554/eLife.73297
- Primary Citation of Related Structures:
6WXH, 6WXI - PubMed Abstract:
The double membrane architecture of Gram-negative bacteria forms a barrier that is impermeable to most extracellular threats. Bacteriocin proteins evolved to exploit the accessible, surface-exposed proteins embedded in the outer membrane to deliver cytotoxic cargo. Colicin E1 is a bacteriocin produced by, and lethal to, Escherichia coli that hijacks the outer membrane proteins (OMPs) TolC and BtuB to enter the cell. Here, we capture the colicin E1 translocation domain inside its membrane receptor, TolC, by high-resolution cryo-electron microscopy to obtain the first reported structure of a bacteriocin bound to TolC. Colicin E1 binds stably to TolC as an open hinge through the TolC pore-an architectural rearrangement from colicin E1's unbound conformation. This binding is stable in live E. coli cells as indicated by single-molecule fluorescence microscopy. Finally, colicin E1 fragments binding to TolC plug the channel, inhibiting its native efflux function as an antibiotic efflux pump, and heightening susceptibility to three antibiotic classes. In addition to demonstrating that these protein fragments are useful starting points for developing novel antibiotic potentiators, this method could be expanded to other colicins to inhibit other OMP functions.
- Center for Computational Biology, The University of Kansas, Lawrence, United States.
Organizational Affiliation: