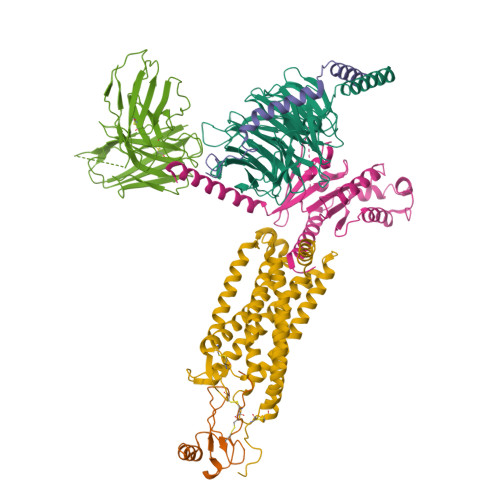
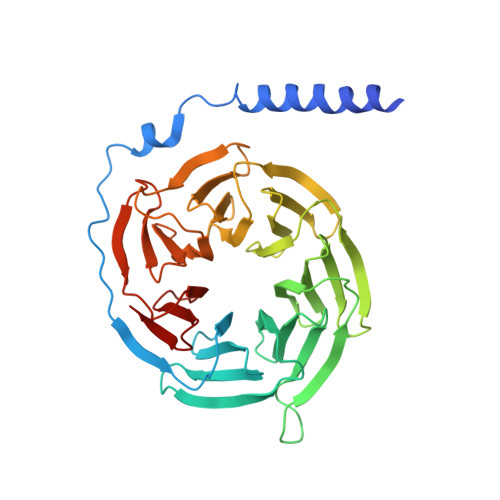
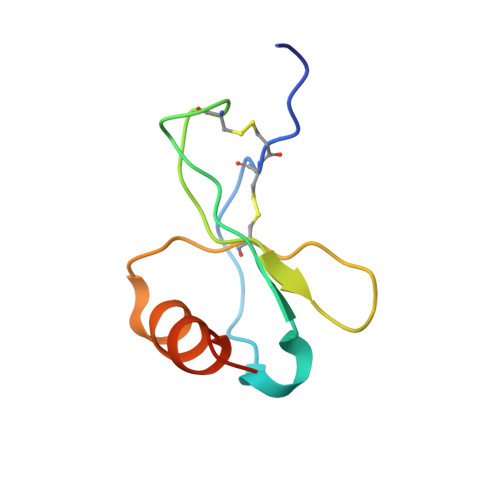
Structural basis for chemokine receptor CCR6 activation by the endogenous protein ligand CCL20.
Wasilko, D.J., Johnson, Z.L., Ammirati, M., Che, Y., Griffor, M.C., Han, S., Wu, H.(2020) Nat Commun 11: 3031-3031
- PubMed: 32541785
- DOI: https://doi.org/10.1038/s41467-020-16820-6
- Primary Citation of Related Structures:
6WWZ - PubMed Abstract:
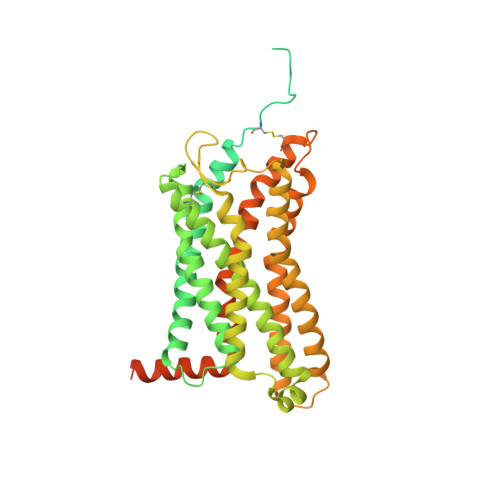
Chemokines are important protein-signaling molecules that regulate various immune responses by activating chemokine receptors which belong to the G protein-coupled receptor (GPCR) superfamily. Despite the substantial progression of our structural understanding of GPCR activation by small molecule and peptide agonists, the molecular mechanism of GPCR activation by protein agonists remains unclear. Here, we present a 3.3-Å cryo-electron microscopy structure of the human chemokine receptor CCR6 bound to its endogenous ligand CCL20 and an engineered Go. CCL20 binds in a shallow extracellular pocket, making limited contact with the core 7-transmembrane (TM) bundle. The structure suggests that this mode of binding induces allosterically a rearrangement of a noncanonical toggle switch and the opening of the intracellular crevice for G protein coupling. Our results demonstrate that GPCR activation by a protein agonist does not always require substantial interactions between ligand and the 7TM core region.
Organizational Affiliation:
Discovery Sciences, Medicine Design, Pfizer Worldwide Research and Development, Groton, CT, 06340, USA.