Crystal structure of PRL phosphatase C104D mutant in complex with the Bateman domain of CNNM magnesium transporter
Kozlov, G., Funato, Y., Chen, S., Zhang, Z., Illes, K., Miki, H., Gehring, K.(2020) J Biol Chem
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2020) J Biol Chem
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein tyrosine phosphatase type IVA 2 | 189 | Homo sapiens | Mutation(s): 1 Gene Names: PTP4A2, PRL2, PTPCAAX2, BM-008 EC: 3.1.3.48 | 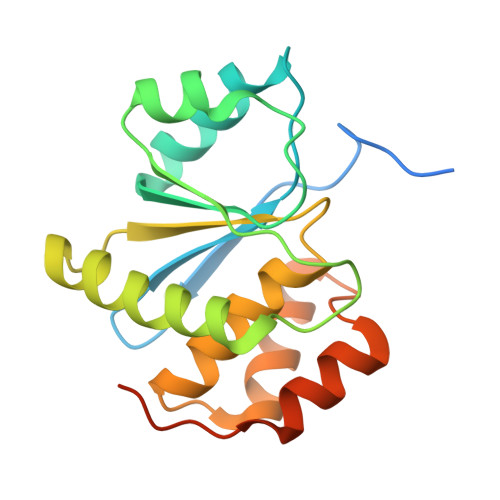 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q12974 (Homo sapiens) Explore Q12974 Go to UniProtKB: Q12974 | |||||
PHAROS: Q12974 GTEx: ENSG00000184007 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q12974 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Metal transporter CNNM3 | 155 | Homo sapiens | Mutation(s): 0 Gene Names: CNNM3, ACDP3 | 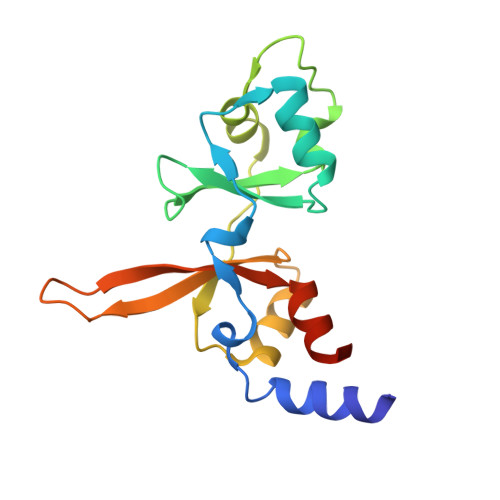 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8NE01 (Homo sapiens) Explore Q8NE01 Go to UniProtKB: Q8NE01 | |||||
PHAROS: Q8NE01 GTEx: ENSG00000168763 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8NE01 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NA Query on NA | C [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 56.857 | α = 90 |
| b = 124.544 | β = 90 |
| c = 153.413 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Natural Sciences and Engineering Research Council (NSERC, Canada) | Canada | RPGIN2014-04686 |