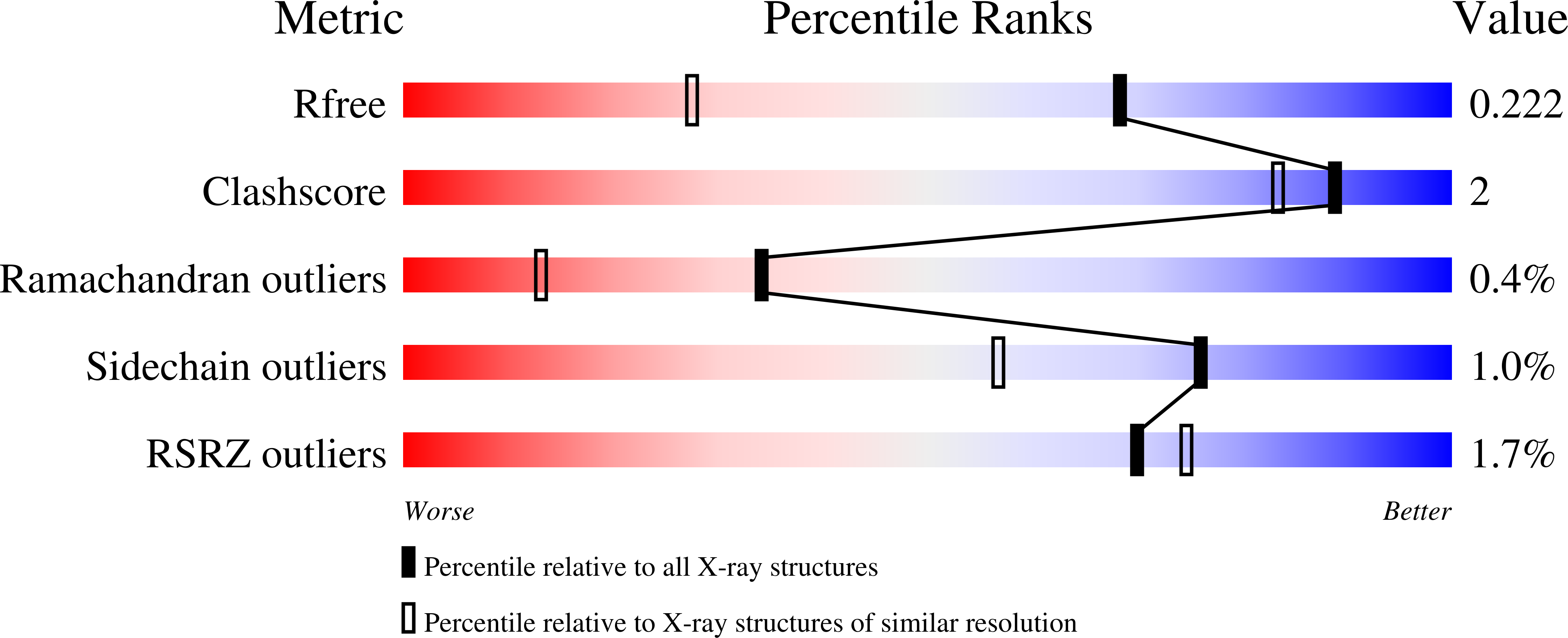
Altered conformational sampling along an evolutionary trajectory changes the catalytic activity of an enzyme.
Kaczmarski, J.A., Mahawaththa, M.C., Feintuch, A., Clifton, B.E., Adams, L.A., Goldfarb, D., Otting, G., Jackson, C.J.(2020) Nat Commun 11: 5945-5945
- PubMed: 33230119
- DOI: https://doi.org/10.1038/s41467-020-19695-9
- Primary Citation of Related Structures:
6WUP - PubMed Abstract:
Several enzymes are known to have evolved from non-catalytic proteins such as solute-binding proteins (SBPs). Although attention has been focused on how a binding site can evolve to become catalytic, an equally important question is: how do the structural dynamics of a binding protein change as it becomes an efficient enzyme? Here we performed a variety of experiments, including propargyl-DO3A-Gd(III) tagging and double electron-electron resonance (DEER) to study the rigid body protein dynamics of reconstructed evolutionary intermediates to determine how the conformational sampling of a protein changes along an evolutionary trajectory linking an arginine SBP to a cyclohexadienyl dehydratase (CDT). We observed that primitive dehydratases predominantly populate catalytically unproductive conformations that are vestiges of their ancestral SBP function. Non-productive conformational states, including a wide-open state, are frozen out of the conformational landscape via remote mutations, eventually leading to extant CDT that exclusively samples catalytically relevant compact states. These results show that remote mutations can reshape the global conformational landscape of an enzyme as a mechanism for increasing catalytic activity.
Organizational Affiliation:
Research School of Chemistry, The Australian National University, Canberra, ACT, 2601, Australia.