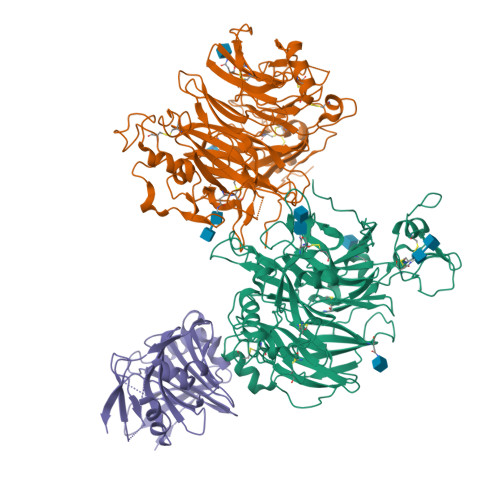
Recognition of Semaphorin Proteins by P. sordellii Lethal Toxin Reveals Principles of Receptor Specificity in Clostridial Toxins.
Lee, H., Beilhartz, G.L., Kucharska, I., Raman, S., Cui, H., Lam, M.H.Y., Liang, H., Rubinstein, J.L., Schramek, D., Julien, J.P., Melnyk, R.A., Taipale, M.(2020) Cell 182: 345
- PubMed: 32589945
- DOI: https://doi.org/10.1016/j.cell.2020.06.005
- Primary Citation of Related Structures:
6WTS - PubMed Abstract:
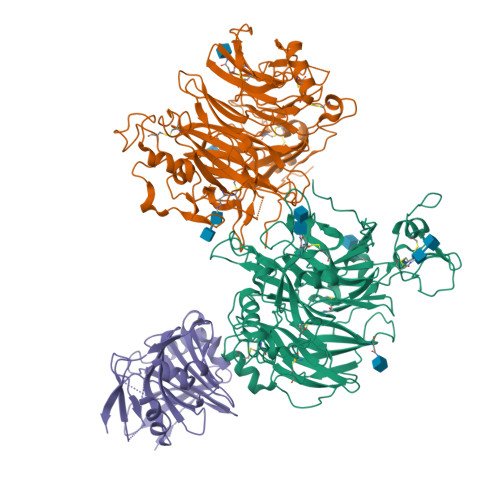
Pathogenic clostridial species secrete potent toxins that induce severe host tissue damage. Paeniclostridium sordellii lethal toxin (TcsL) causes an almost invariably lethal toxic shock syndrome associated with gynecological infections. TcsL is 87% similar to C. difficile TcdB, which enters host cells via Frizzled receptors in colon epithelium. However, P. sordellii infections target vascular endothelium, suggesting that TcsL exploits another receptor. Here, using CRISPR/Cas9 screening, we establish semaphorins SEMA6A and SEMA6B as TcsL receptors. We demonstrate that recombinant SEMA6A can protect mice from TcsL-induced edema. A 3.3 Å cryo-EM structure shows that TcsL binds SEMA6A with the same region that in TcdB binds structurally unrelated Frizzled. Remarkably, 15 mutations in this evolutionarily divergent surface are sufficient to switch binding specificity of TcsL to that of TcdB. Our findings establish semaphorins as physiologically relevant receptors for TcsL and reveal the molecular basis for the difference in tissue targeting and disease pathogenesis between highly related toxins.
Organizational Affiliation:
Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, ON M5S 3E1, Canada.