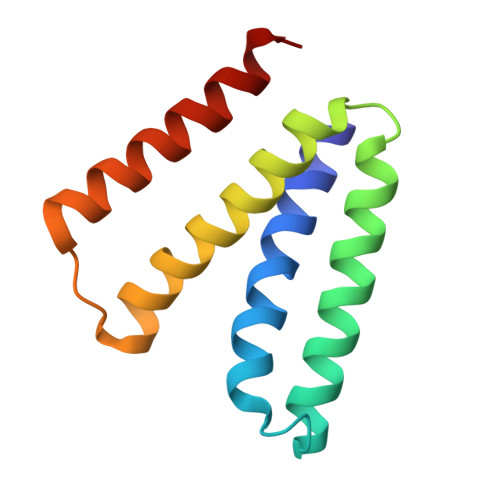
The structural basis of promiscuity in small multidrug resistance transporters.
Kermani, A.A., Macdonald, C.B., Burata, O.E., Ben Koff, B., Koide, A., Denbaum, E., Koide, S., Stockbridge, R.B.(2020) Nat Commun 11: 6064-6064
- PubMed: 33247110
- DOI: https://doi.org/10.1038/s41467-020-19820-8
- Primary Citation of Related Structures:
6WK5, 6WK8, 6WK9 - PubMed Abstract:
By providing broad resistance to environmental biocides, transporters from the small multidrug resistance (SMR) family drive the spread of multidrug resistance cassettes among bacterial populations. A fundamental understanding of substrate selectivity by SMR transporters is needed to identify the types of selective pressures that contribute to this process. Using solid-supported membrane electrophysiology, we find that promiscuous transport of hydrophobic substituted cations is a general feature of SMR transporters. To understand the molecular basis for promiscuity, we solved X-ray crystal structures of a SMR transporter Gdx-Clo in complex with substrates to a maximum resolution of 2.3 Å. These structures confirm the family's extremely rare dual topology architecture and reveal a cleft between two helices that provides accommodation in the membrane for the hydrophobic substituents of transported drug-like cations.
Organizational Affiliation:
Department of Molecular, Cellular, and Developmental Biology, University of Michigan, Ann Arbor, MI, 48109, USA.