FMN riboswitch aptamer symmetry facilitates conformational switching through mutually exclusive coaxial stacking configurations.
Wilt, H.M., Yu, P., Tan, K., Wang, Y.X., Stagno, J.R.(2020) J Struct Biol X 4: 100035-100035
- PubMed: 33103111
- DOI: https://doi.org/10.1016/j.yjsbx.2020.100035
- Primary Citation of Related Structures:
6WJR, 6WJS - PubMed Abstract:
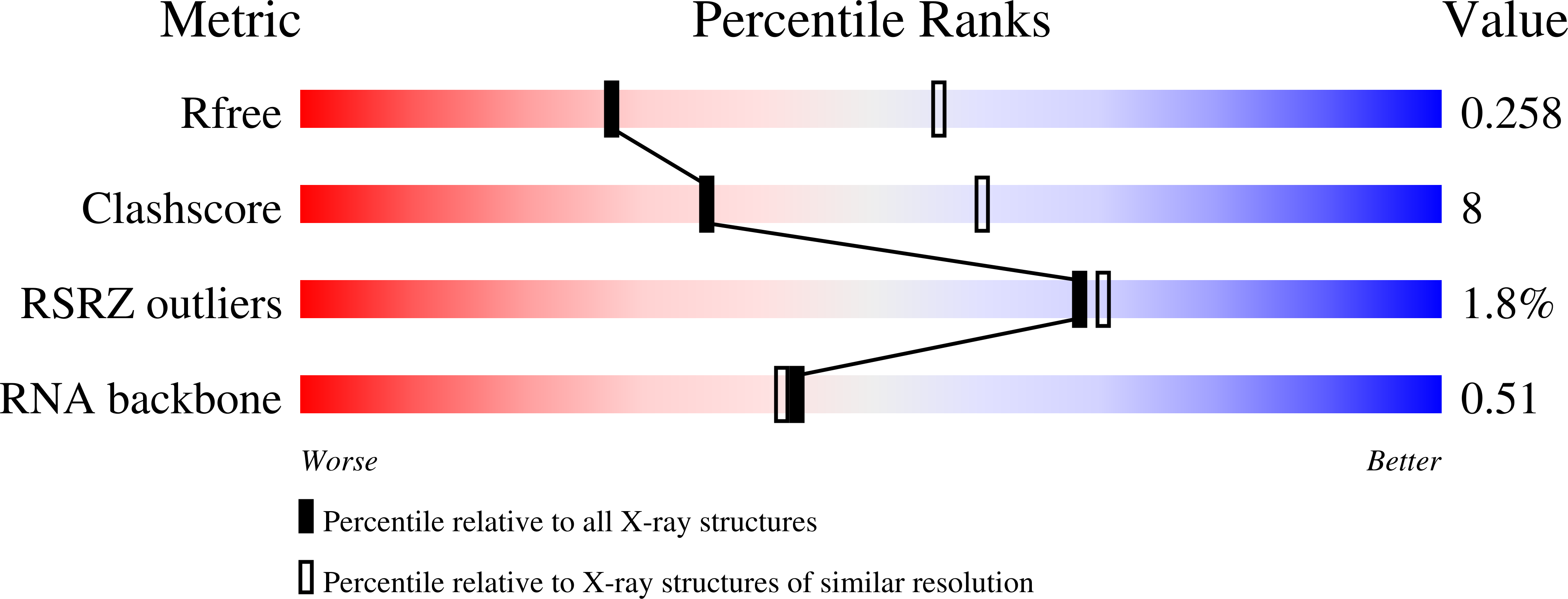
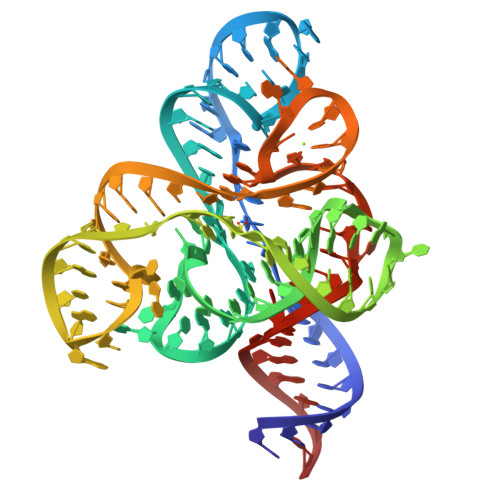
Knowledge of both apo and holo states of riboswitches aid in elucidating the various mechanisms of ligand-induced conformational "switching" that underpin their gene-regulating capabilities. Previous structural studies on the flavin mononucleotide (FMN)-binding aptamer of the FMN riboswitch, however, have revealed minimal conformational changes associated with ligand binding that do not adequately explain the basis for the switching behavior. We have determined a 2.7-Å resolution crystal structure of the ligand-free FMN riboswitch aptamer that is distinct from previously reported structures, particularly in the conformation and orientation of the P1 and P4 helices. The nearly symmetrical tertiary structure provides a mechanism by which one of two pairs of adjacent helices (P3/P4 or P1/P6) undergo collinear stacking in a mutually exclusive manner, in the absence or presence of ligand, respectively. Comparison of these structures suggests the stem-loop that includes P4 and L4 is important for maintaining a global conformational state that, in the absence of ligand, disfavors formation of the P1 regulatory helix. Together, these results provide further insight to the structural basis for conformational switching of the FMN riboswitch.
Organizational Affiliation:
Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.