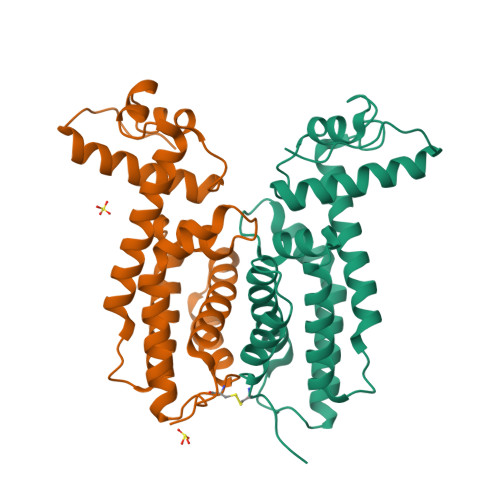
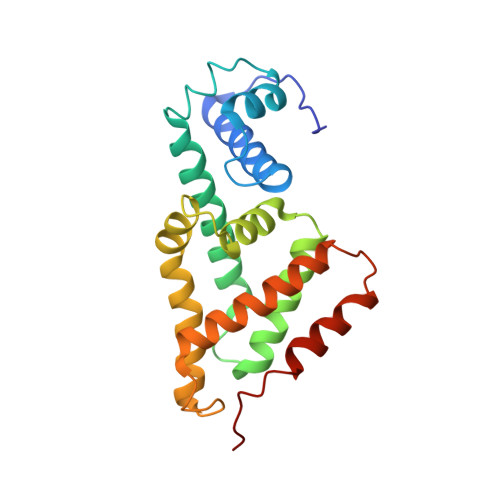
The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Newman, J.D., Russell, M.M., Fan, L., Wang, Y.X., Gonzalez-Gutierrez, G., van Kessel, J.C.(2021) Nucleic Acids Res 49: 5967-5984
- PubMed: 34023896
- DOI: https://doi.org/10.1093/nar/gkab387
- Primary Citation of Related Structures:
6WAE, 6WAF, 6WAG, 6WAH, 6WAI - PubMed Abstract:
Quorum sensing gene expression in vibrios is regulated by the LuxR/HapR family of transcriptional factors, which includes Vibrio vulnificus SmcR. The consensus binding site of Vibrio LuxR/HapR/SmcR proteins is palindromic but highly degenerate with sequence variations at each promoter. To examine the mechanism by which SmcR recognizes diverse DNA sites, we generated SmcR separation-of-function mutants that either repress or activate transcription but not both. SmcR N55I is restricted in recognition of single base-pair variations in DNA binding site sequences and thus is defective at transcription activation but retains interaction with RNA polymerase (RNAP) alpha. SmcR S76A, L139R and N142D substitutions disrupt the interaction with RNAP alpha but retain functional DNA binding activity. X-ray crystallography and small angle X-ray scattering data show that the SmcR DNA binding domain exists in two conformations (wide and narrow), and the protein complex forms a mixture of dimers and tetramers in solution. The three RNAP interaction-deficient variants also have two DNA binding domain conformations, whereas SmcR N55I exhibits only the wide conformation. These data support a model in which two mechanisms drive SmcR transcriptional activation: interaction with RNAP and a multi-conformational DNA binding domain that permits recognition of variable DNA sites.
Organizational Affiliation:
Department of Biology, Indiana University, 1001 E 3rd St, Bloomington, IN 47405, USA.