Development of Antibiotics That Dysregulate the Neisserial ClpP Protease.
Binepal, G., Mabanglo, M.F., Goodreid, J.D., Leung, E., Barghash, M.M., Wong, K.S., Lin, F., Cossette, M., Bansagi, J., Song, B., Balasco Serrao, V.H., Pai, E.F., Batey, R.A., Gray-Owen, S.D., Houry, W.A.(2020) ACS Infect Dis 6: 3224-3236
- PubMed: 33237740
- DOI: https://doi.org/10.1021/acsinfecdis.0c00599
- Primary Citation of Related Structures:
6W9T - PubMed Abstract:
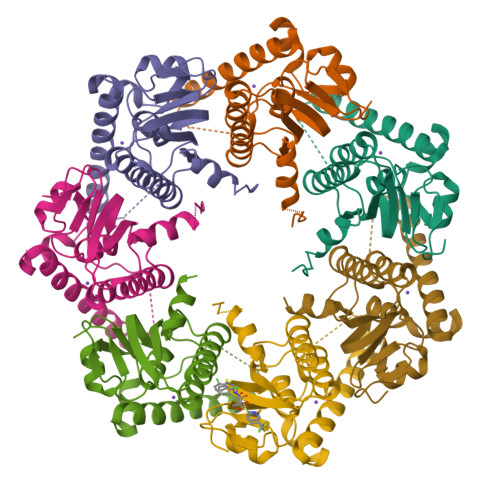
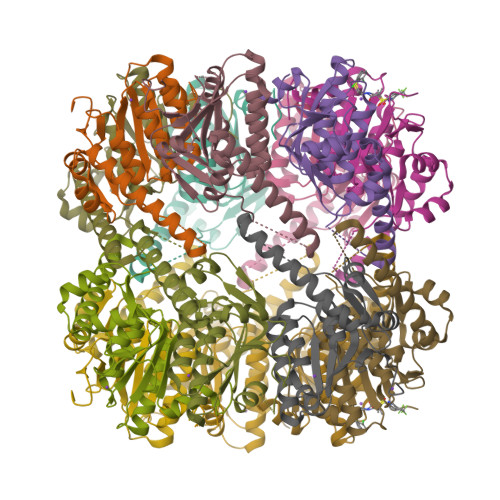
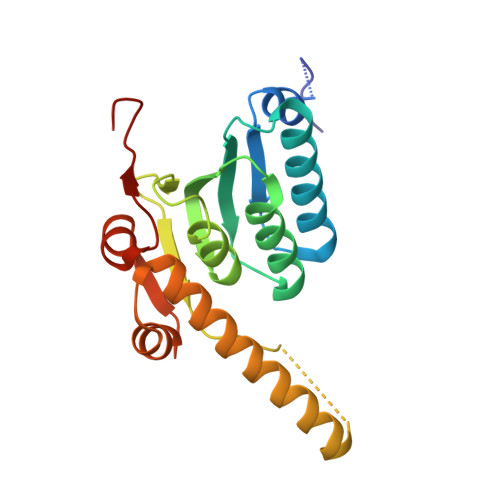
Evolving antimicrobial resistance has motivated the search for novel targets and alternative therapies. Caseinolytic protease (ClpP) has emerged as an enticing new target since its function is conserved and essential for bacterial fitness, and because its inhibition or dysregulation leads to bacterial cell death. ClpP protease function controls global protein homeostasis and is, therefore, crucial for the maintenance of the bacterial proteome during growth and infection. Previously, acyldepsipeptides (ADEPs) were discovered to dysregulate ClpP, leading to bactericidal activity against both actively growing and dormant Gram-positive pathogens. Unfortunately, these compounds had very low efficacy against Gram-negative bacteria. Hence, we sought to develop non-ADEP ClpP-targeting compounds with activity against Gram-negative species and called these activators of self-compartmentalizing proteases (ACPs). These ACPs bind and dysregulate ClpP in a manner similar to ADEPs, effectively digesting bacteria from the inside out. Here, we performed further ACP derivatization and testing to improve the efficacy and breadth of coverage of selected ACPs against Gram-negative bacteria. We observed that a diverse collection of Neisseria meningitidis and Neisseria gonorrhoeae clinical isolates were exquisitely sensitive to these ACP analogues. Furthermore, based on the ACP-ClpP cocrystal structure solved here, we demonstrate that ACPs could be designed to be species specific. This validates the feasibility of drug-based targeting of ClpP in Gram-negative bacteria.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, Toronto, Ontario M5G 1M1, Canada.