Structural basis for Ca2+-dependent activation of a plant metacaspase.
Zhu, P., Yu, X.H., Wang, C., Zhang, Q., Liu, W., McSweeney, S., Shanklin, J., Lam, E., Liu, Q.(2020) Nat Commun 11: 2249-2249
- PubMed: 32382010
- DOI: https://doi.org/10.1038/s41467-020-15830-8
- Primary Citation of Related Structures:
6W8R, 6W8S, 6W8T - PubMed Abstract:
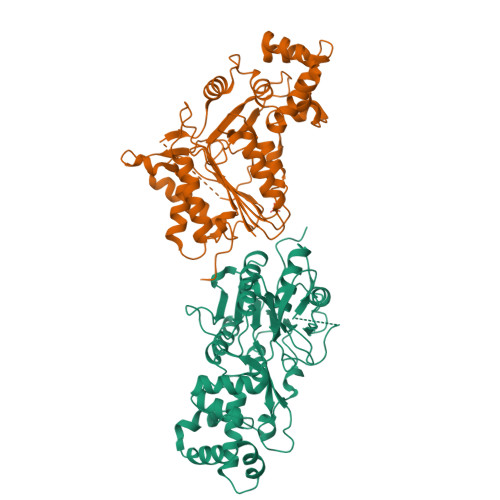
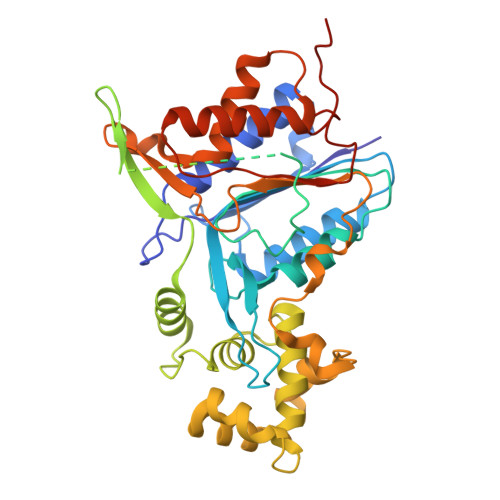
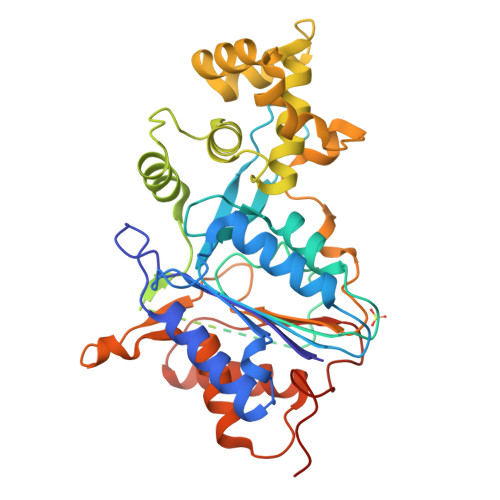
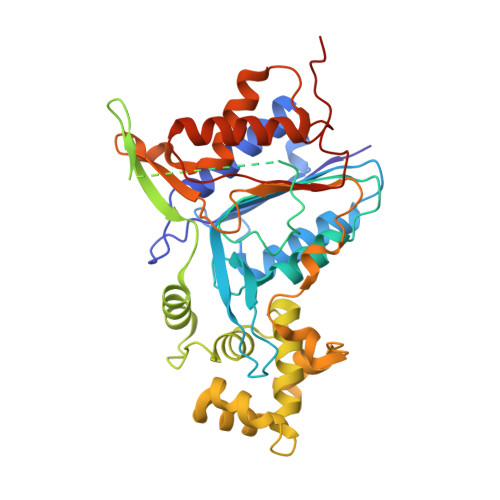
Plant metacaspases mediate programmed cell death in development, biotic and abiotic stresses, damage-induced immune response, and resistance to pathogen attack. Most metacaspases require Ca 2+ for their activation and substrate processing. However, the Ca 2+ -dependent activation mechanism remains elusive. Here we report the crystal structures of Metacaspase 4 from Arabidopsis thaliana (AtMC4) that modulates Ca 2+ -dependent, damage-induced plant immune defense. The AtMC4 structure exhibits an inhibitory conformation in which a large linker domain blocks activation and substrate access. In addition, the side chain of Lys225 in the linker domain blocks the active site by sitting directly between two catalytic residues. We show that the activation of AtMC4 and cleavage of its physiological substrate involve multiple cleavages in the linker domain upon activation by Ca 2+ . Our analysis provides insight into the Ca 2+ -dependent activation of AtMC4 and lays the basis for tuning its activity in response to stresses for engineering of more sustainable crops for food and biofuels.
Organizational Affiliation:
Biology Department, Brookhaven National Laboratory, Upton, NY, USA.