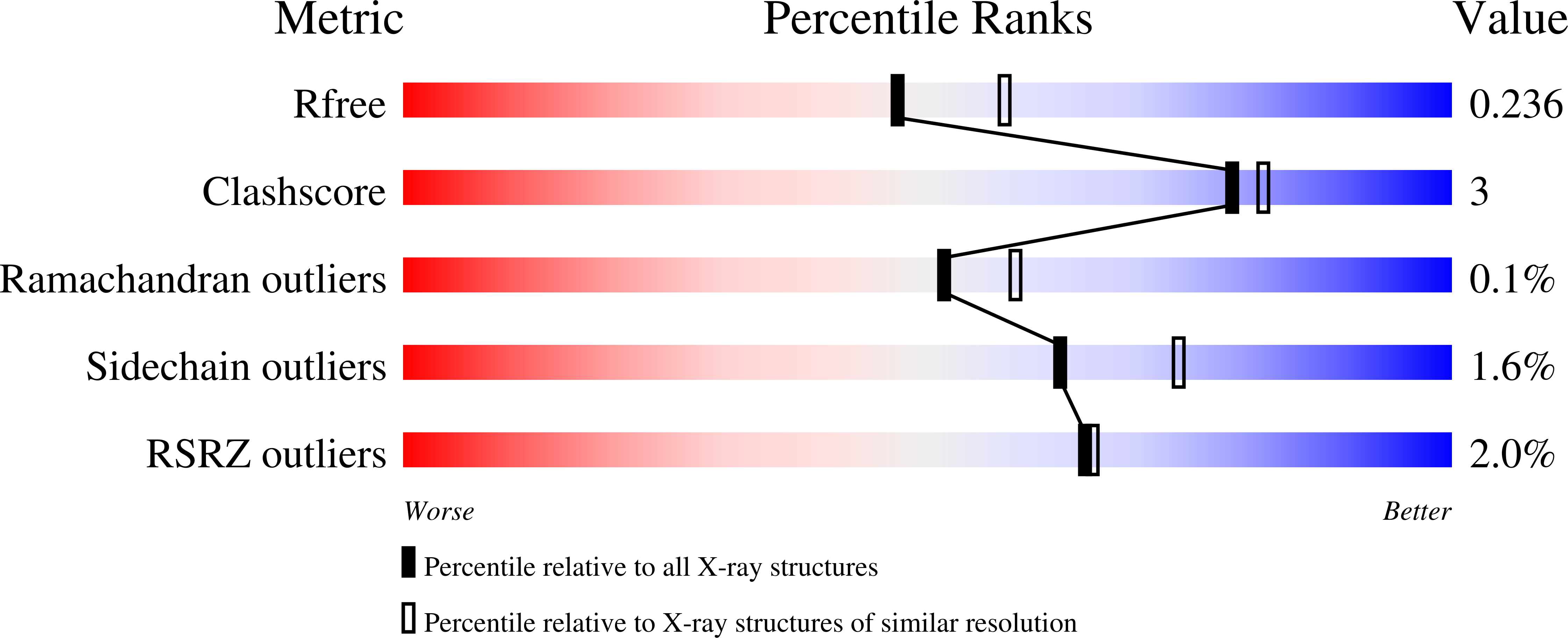
Structural Characterization of BTK:PROTAC:cIAP Ternary Complexes: From Snapshots to Ensembles
Calabrese, M.F., Schiemer, J.S., Horst, R., Meng, Y., Montgomery, J., Xu, Y., Feng, X., Borzilleri, K., Uccello, D.P., Leverett, C., Brown, S., Che, Y., Brown, M.F., Hayward, M.M., Gilbert, A.M., Noe, M.C.(2020) Nat Chem Biol