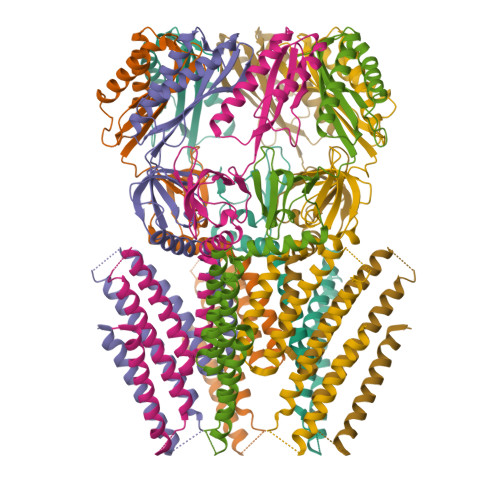
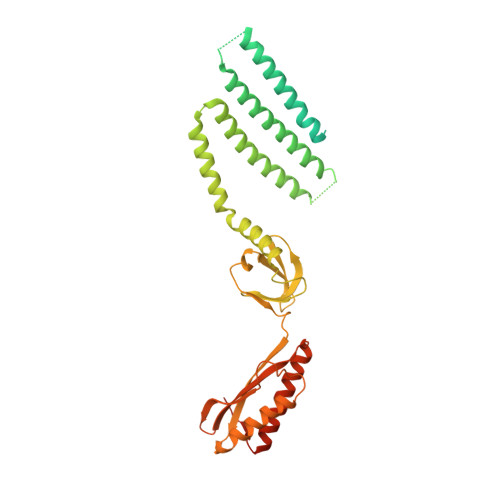
Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Deng, Z., Maksaev, G., Schlegel, A.M., Zhang, J., Rau, M., Fitzpatrick, J.A.J., Haswell, E.S., Yuan, P.(2020) Nat Commun 11: 3690-3690
- PubMed: 32704140
- DOI: https://doi.org/10.1038/s41467-020-17538-1
- Primary Citation of Related Structures:
6VXM, 6VXN, 6VXP - PubMed Abstract:
Mechanosensitive ion channels transduce physical force into electrochemical signaling that underlies an array of fundamental physiological processes, including hearing, touch, proprioception, osmoregulation, and morphogenesis. The mechanosensitive channels of small conductance (MscS) constitute a remarkably diverse superfamily of channels critical for management of osmotic pressure. Here, we present cryo-electron microscopy structures of a MscS homolog from Arabidopsis thaliana, MSL1, presumably in both the closed and open states. The heptameric MSL1 channel contains an unusual bowl-shaped transmembrane region, which is reminiscent of the evolutionarily and architecturally unrelated mechanosensitive Piezo channels. Upon channel opening, the curved transmembrane domain of MSL1 flattens and expands. Our structures, in combination with functional analyses, delineate a structural mechanism by which mechanosensitive channels open under increased membrane tension. Further, the shared structural feature between unrelated channels suggests the possibility of a unified mechanical gating mechanism stemming from membrane deformation induced by a non-planar transmembrane domain.
Organizational Affiliation:
Department of Cell Biology and Physiology, Washington University School of Medicine, Saint Louis, MO, 63110, USA.