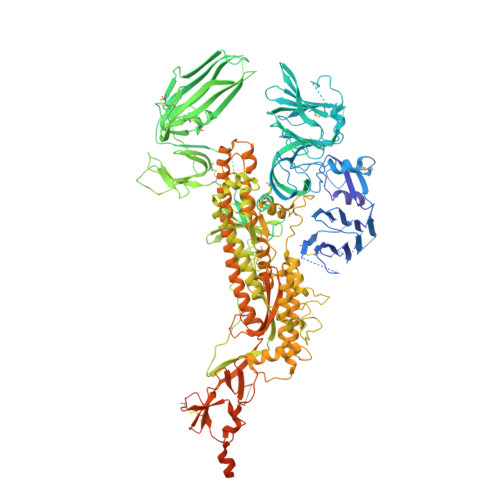
Structure and immune recognition of the porcine epidemic diarrhea virus spike protein.
Kirchdoerfer, R.N., Bhandari, M., Martini, O., Sewall, L.M., Bangaru, S., Yoon, K.J., Ward, A.B.(2021) Structure 29: 385-392.e5
- PubMed: 33378641
- DOI: https://doi.org/10.1016/j.str.2020.12.003
- Primary Citation of Related Structures:
6VV5 - PubMed Abstract:
Porcine epidemic diarrhea virus (PEDV) is an alphacoronavirus responsible for significant morbidity and mortality in pigs. A key determinant of viral tropism and entry, the PEDV spike protein is a key target for the host antibody response and a good candidate for a protein-based vaccine immunogen. We used electron microscopy to evaluate the PEDV spike structure, as well as pig polyclonal antibody responses to viral infection. The structure of the PEDV spike reveals a configuration similar to that of HuCoV-NL63. Several PEDV protein-protein interfaces are mediated by non-protein components, including a glycan at Asn264 and two bound palmitoleic acid molecules. The polyclonal antibody response to PEDV infection shows a dominance of epitopes in the S1 region. This structural and immune characterization provides insights into coronavirus spike stability determinants and explores the immune landscape of viral spike proteins.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.