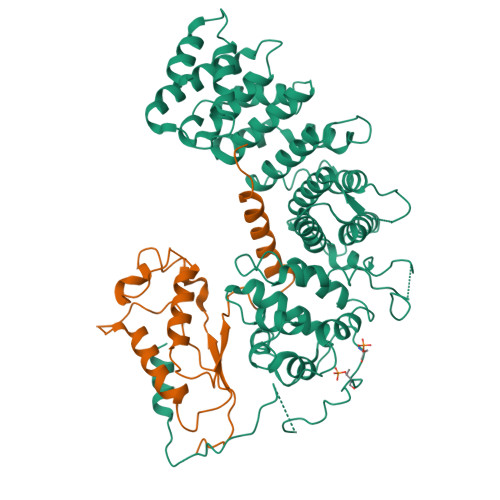
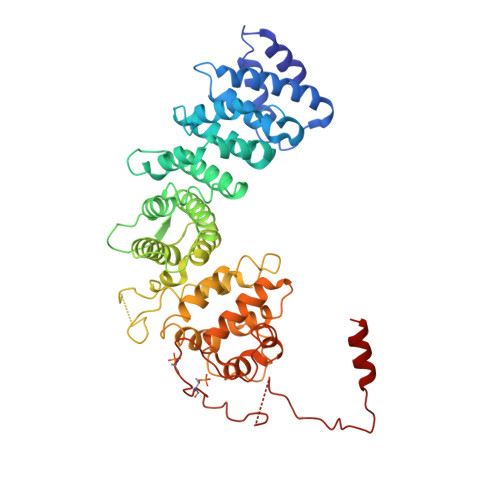
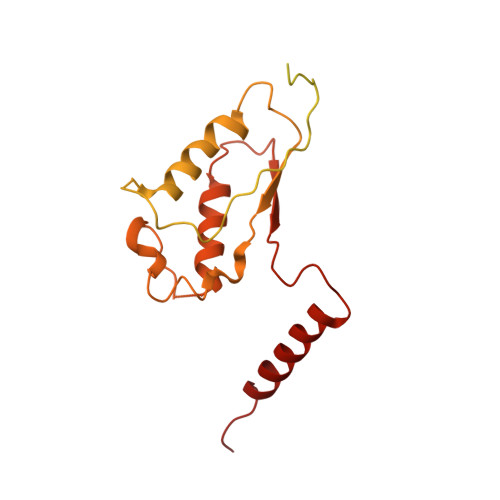
Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Seven, A.B., Hilger, D., Papasergi-Scott, M.M., Zhang, L., Qu, Q., Kobilka, B.K., Tall, G.G., Skiniotis, G.(2020) Cell Rep 30: 3699-3709.e6
- PubMed: 32126208
- DOI: https://doi.org/10.1016/j.celrep.2020.02.086
- Primary Citation of Related Structures:
6VU5, 6VU8 - PubMed Abstract:
Many chaperones promote nascent polypeptide folding followed by substrate release through ATP-dependent conformational changes. Here we show cryoEM structures of Gα subunit folding intermediates in complex with full-length Ric-8A, a unique chaperone-client system in which substrate release is facilitated by guanine nucleotide binding to the client G protein. The structures of Ric-8A-Gα i and Ric-8A-Gα q complexes reveal that the chaperone employs its extended C-terminal region to cradle the Ras-like domain of Gα, positioning the Ras core in contact with the Ric-8A core while engaging its switch2 nucleotide binding region. The C-terminal α5 helix of Gα is held away from the Ras-like domain through Ric-8A core domain interactions, which critically depend on recognition of the Gα C terminus by the chaperone. The structures, complemented with biochemical and cellular chaperoning data, support a folding quality control mechanism that ensures proper formation of the C-terminal α5 helix before allowing GTP-gated release of Gα from Ric-8A.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA 94305, USA; Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.