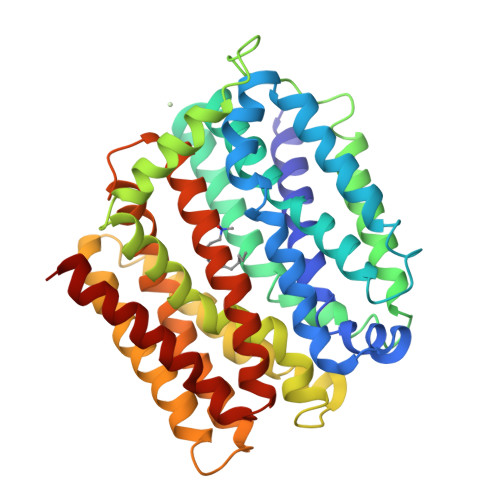
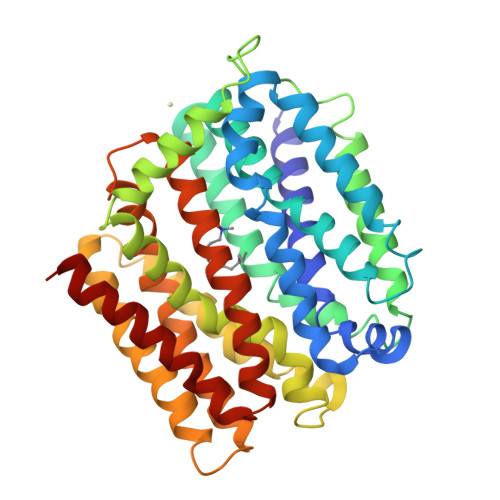
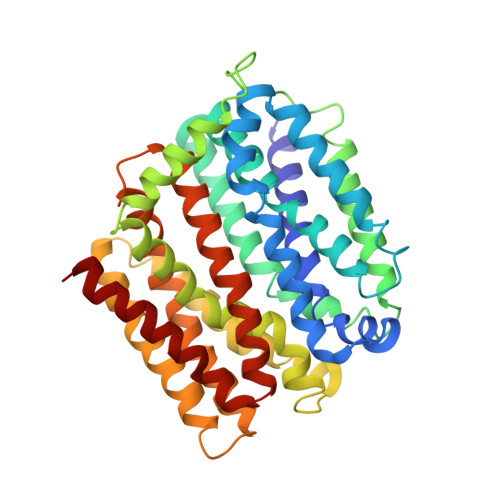
Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Wu, H.H., Symersky, J., Lu, M.(2020) Sci Rep 10: 3949-3949
- PubMed: 32127561
- DOI: https://doi.org/10.1038/s41598-020-60332-8
- Primary Citation of Related Structures:
6VRZ, 6VS0, 6VS1, 6VS2 - PubMed Abstract:
The rapid increase of multidrug resistance poses urgent threats to human health. Multidrug transporters prompt multidrug resistance by exporting different therapeutics across cell membranes, often by utilizing the H + electrochemical gradient. MdfA from Escherichia coli is a prototypical H + -dependent multidrug transporter belonging to the Major Facilitator Superfamily. Prior studies revealed unusual flexibility in the coupling between multidrug binding and deprotonation in MdfA, but the mechanistic basis for this flexibility was obscure. Here we report the X-ray structures of a MdfA mutant E26T/D34M/A150E, wherein the multidrug-binding and protonation sites were revamped, separately bound to three different substrates at resolutions up to 2.0 Å. To validate the functional relevance of these structures, we conducted mutational and biochemical studies. Our data elucidated intermediate states during antibiotic recognition and suggested structural changes that accompany the substrate-evoked deprotonation of E26T/D34M/A150E. These findings help to explain the mechanistic flexibility in drug/H + coupling observed in MdfA and may inspire therapeutic development to preempt efflux-mediated antimicrobial resistance.
Organizational Affiliation:
Center for Proteomics and Molecular Therapeutics, Rosalind Franklin University of Medicine and Science, 3333 Green Bay Road, North Chicago, IL, 60064, USA.