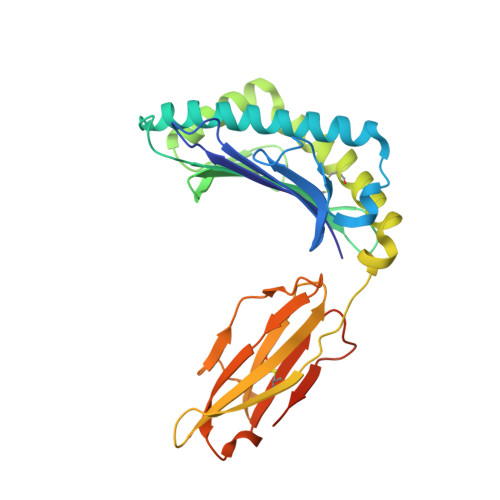
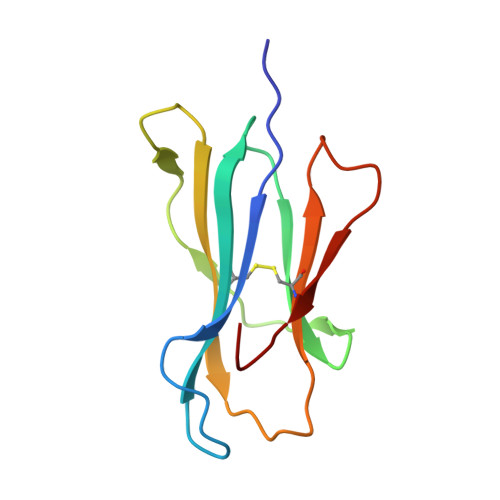
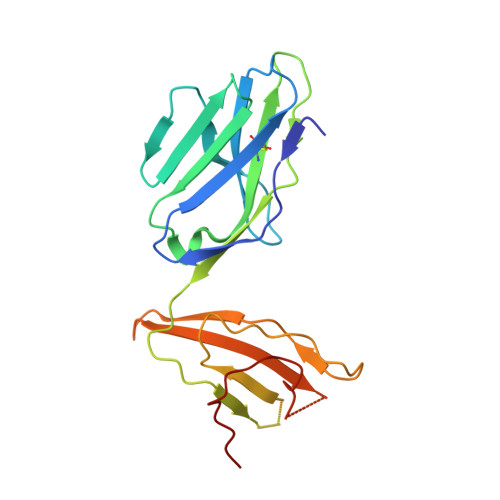
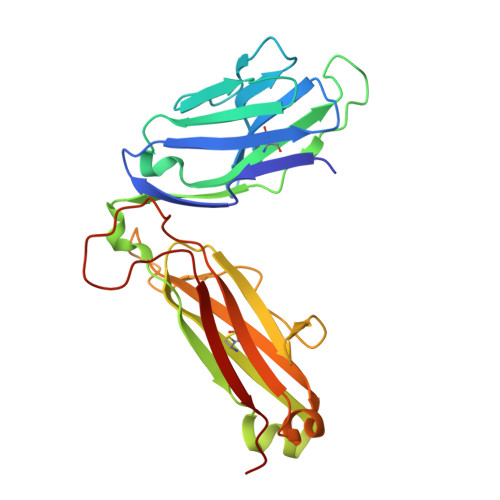
Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Wu, D., Gallagher, D.T., Gowthaman, R., Pierce, B.G., Mariuzza, R.A.(2020) Nat Commun 11: 2908-2908
- PubMed: 32518267
- DOI: https://doi.org/10.1038/s41467-020-16755-y
- Primary Citation of Related Structures:
6VQO, 6VR1, 6VR5, 6VRM, 6VRN, 6VTC, 6VTH - PubMed Abstract:
Adoptive cell therapy (ACT) with tumor-specific T cells can mediate cancer regression. The main target of tumor-specific T cells are neoantigens arising from mutations in self-proteins. Although the majority of cancer neoantigens are unique to each patient, and therefore not broadly useful for ACT, some are shared. We studied oligoclonal T-cell receptors (TCRs) that recognize a shared neoepitope arising from a driver mutation in the p53 oncogene (p53R175H) presented by HLA-A2. Here we report structures of wild-type and mutant p53-HLA-A2 ligands, as well as structures of three tumor-specific TCRs bound to p53R175H-HLA-A2. These structures reveal how a driver mutation in p53 rendered a self-peptide visible to T cells. The TCRs employ structurally distinct strategies that are highly focused on the mutation to discriminate between mutant and wild-type p53. The TCR-p53R175H-HLA-A2 complexes provide a framework for designing TCRs to improve potency for ACT without sacrificing specificity.
Organizational Affiliation:
W.M. Keck Laboratory for Structural Biology, University of Maryland Institute for Bioscience and Biotechnology Research, Rockville, MD, 20850, USA.